mirror of
https://github.com/qurator-spk/eynollah.git
synced 2026-02-21 00:41:56 +01:00
Merge remote-tracking branch 'origin/main' into basic-python-tooling
This commit is contained in:
commit
f210054eb4
2 changed files with 215 additions and 284 deletions
80
README.md
80
README.md
|
|
@ -1,18 +1,35 @@
|
|||
# Textline Detection
|
||||
> Detect textlines in document images
|
||||
**WARNING! This tool is currently** 
|
||||
**For any planned changes, please have a look at the [Pull Requests](https://github.com/qurator-spk/eynollah/pulls).**
|
||||
|
||||
# Eynollah
|
||||
> Document Layout Analysis
|
||||
|
||||
## Introduction
|
||||
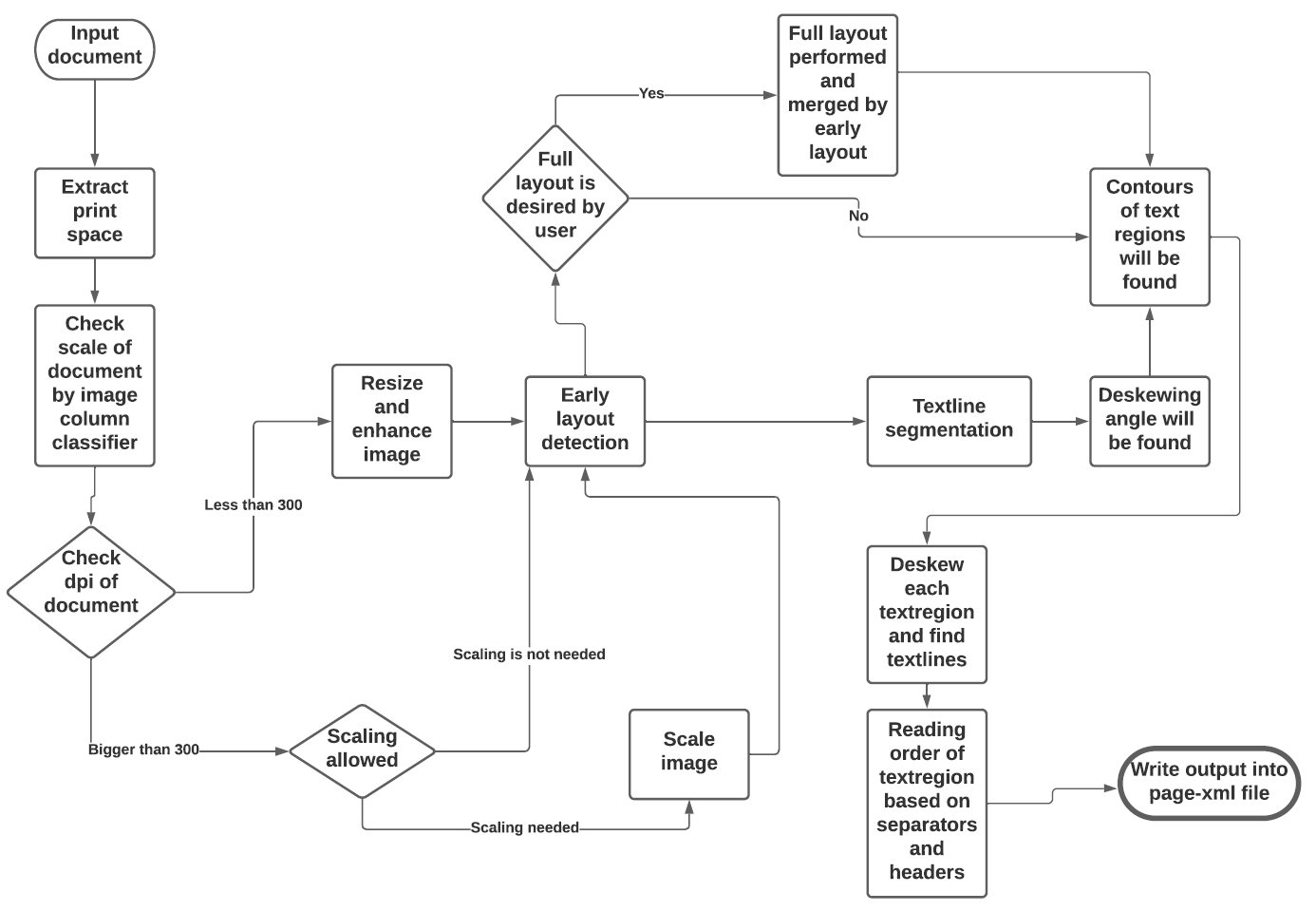
This tool (eynollah) performs border, region and textline detection and scaling and enhancing from document image data and returns the results as [PAGE-XML](https://github.com/PRImA-Research-Lab/PAGE-XML).
|
||||
The goal of this project is to extract textlines of a document in order to feed them to an OCR model. This is achieved by four successive stages as follows:
|
||||
* [Border detection](https://github.com/qurator-spk#border-detection)
|
||||
* [Layout detection](https://github.com/qurator-spk#layout-detection)
|
||||
* [Textline detection](https://github.com/qurator-spk#textline-detection)
|
||||
* [Image enhancement](https://github.com/qurator-spk#Image_enhancement)
|
||||
* [Scale classification](https://github.com/qurator-spk#Scale_classification)
|
||||
* [Heuristic methods](https://github.com/qurator-spk#heuristic-methods)
|
||||
This tool performs document layout analysis (segmentation) from image data and returns the results as [PAGE-XML](https://github.com/PRImA-Research-Lab/PAGE-XML).
|
||||
|
||||
It can currently detect the following layout classes/elements:
|
||||
* Border
|
||||
* Textregion
|
||||
* Image
|
||||
* Textline
|
||||
* Separator
|
||||
* Marginalia
|
||||
* Initial (Drop Capital)
|
||||
|
||||
In addition, the tool can be used to detect the _Reading Order_ of regions. The final goal is to feed the output to an OCR model.
|
||||
|
||||
The tool uses a combination of various models and heuristics (see flowchart below for the different stages and how they interact):
|
||||
* [Border detection](https://github.com/qurator-spk/eynollah#border-detection)
|
||||
* [Layout detection](https://github.com/qurator-spk/eynollah#layout-detection)
|
||||
* [Textline detection](https://github.com/qurator-spk/eynollah#textline-detection)
|
||||
* [Image enhancement](https://github.com/qurator-spk/eynollah#Image_enhancement)
|
||||
* [Scale classification](https://github.com/qurator-spk/eynollah#Scale_classification)
|
||||
* [Heuristic methods](https://https://github.com/qurator-spk/eynollah#heuristic-methods)
|
||||
|
||||
The first three stages are based on [pixelwise segmentation](https://github.com/qurator-spk/sbb_pixelwise_segmentation).
|
||||
|
||||
|
||||
|
||||
## Border detection
|
||||
For the purpose of text recognition (OCR) and in order to avoid noise being introduced from texts outside the printspace, one first needs to detect the border of the printed frame. This is done by a binary pixelwise-segmentation model trained on a dataset of 2,000 documents where about 1,200 of them come from the [dhSegment](https://github.com/dhlab-epfl/dhSegment/) project (you can download the dataset from [here](https://github.com/dhlab-epfl/dhSegment/releases/download/v0.2/pages.zip)) and the remainder having been annotated in SBB. For border detection, the model needs to be fed with the whole image at once rather than separated in patches.
|
||||
|
||||
|
|
@ -38,43 +55,42 @@ Some heuristic methods are also employed to further improve the model prediction
|
|||
* Finally, using the derived coordinates, bounding boxes are determined for each textline.
|
||||
|
||||
## Installation
|
||||
|
||||
```
|
||||
pip install .
|
||||
# or
|
||||
make install
|
||||
```
|
||||
`pip install .`
|
||||
|
||||
### Models
|
||||
|
||||
<!-- TODO add link to models -->
|
||||
|
||||
In order to run this tool you also need trained models. You can download [pretrained models from qurator-data](https://qurator-data.de/).
|
||||
In order to run this tool you also need trained models. You can download our pretrained models from [qurator-data.de](https://qurator-data.de/eynollah/).
|
||||
|
||||
## Usage
|
||||
|
||||
The basic command-line interface can be called like this:
|
||||
|
||||
eynollah -i <image file name> -o <directory to write output xml or enhanced image> -m <directory of models> -fl <if this parameter is set to true, full layout will be done> -ae <if true, this tool would resize and enhance image and result will be written in output> -as <if true, this tool would check whether the document needs scaling or not> -cl <if true, the tool will try to extract contours of texlines instead of rectangle bounding boxes> -si <if a directory is given here, this tool would write image regions inside documents there>
|
||||
eynollah \
|
||||
-i <image file name> \
|
||||
-o <directory to write output xml or enhanced image> \
|
||||
-m <directory of models> \
|
||||
-fl <if true, the tool will perform full layout analysis> \
|
||||
-ae <if true, the tool will resize and enhance the image and produce the resulting image as output> \
|
||||
-as <if true, the tool will check whether the document needs rescaling or not> \
|
||||
-cl <if true, the tool will try to extract the contours of texlines instead of rectangle bounding boxes> \
|
||||
-si <if a directory is given here, the tool will output image regions inside documents there>
|
||||
|
||||
The tool does accepts and works better on original images (RGB format).
|
||||
The tool does accept and works better on original images (RGB format) than binarized images.
|
||||
|
||||
### How and where to use
|
||||
### How to use
|
||||
|
||||
First of all, for this model we have trained 9 models which are doing different jobs like size detection (or column classifier), enhancing, page extraction, main layout detection, full layout detection and textline detetction. But this does not mean all those 9 models are needed for each document. Based on document and parameters it can be different. It is worthy to mention that with this tool we are able to detect reading order of text regions for simple documents (I will not go in detail with order of reading since it is a complex issue and many factors play a role about it).
|
||||
First of all, this model makes use of up to 9 trained models which are responsible for different operations like size detection, column classification, image enhancement, page extraction, main layout detection, full layout detection and textline detection. But this does not mean that all 9 models are always required for every document. Based on the document characteristics and parameters specified, different scenarios can be applied.
|
||||
|
||||
* If none of parameters is set to true, this tool will try to do a layout detection of main regions (background, text, images, separators and marginals). Actually, advantage of this tool is that it has tried to extract main text regions separately as much as possible.
|
||||
* If none of the parameters is set to `true`, the tool will perform a layout detection of main regions (background, text, images, separators and marginals). An advantage of this tool is that it tries to extract main text regions separately as much as possible.
|
||||
|
||||
* If you set -ae(allow enhancement) paremeter to true, this tool would check first dpi of document and if it is less than 300 then our tool first will resize it and then enhancement will occur. In fact enhancemnet can take place even without this option but by setting this option to true layout (better say xml data) will be written on resized and enhanced image instead of original image.
|
||||
* If you set `-ae` (**a**llow image **e**nhancement) parameter to `true`, the tool will first check the ppi (pixel-per-inch) of the image and when it is less than 300, the tool will resize it and only then image enhancement will occur. Image enhancement can also take place without this option, but by setting this option to `true`, the layout xml data (e.g. coordinates) will be based on the resized and enhanced image instead of the original image.
|
||||
|
||||
* Some documents quality are really good but their scale is extremly big and therefore the performance of tool decreases. In those cases you can set -as (allow scaling) to true. With this option our tool first would try to scale image and then layout detection process will begin.
|
||||
* For some documents, while the quality is good, their scale is extremly large and the performance of tool decreases. In such cases you can set `-as` (**a**llow **s**caling) to `true`. With this option enabled, the tool will try to rescale the image and only then the layout detection process will begin.
|
||||
|
||||
* If you care about drop capitals and headings you can set -fl (full layout) to true. As we can see in the case of full layout we can detect 7 elements of document.
|
||||
* If you care about drop capitals (initials) and headings, you can set `-fl` (**f**ull **l**ayout) to `true`. As we can see in the case of full layout, we can currently distinguish 7 document layout classes/elements.
|
||||
|
||||
* We face documents which include curved header or curved lines and it is abvious that a rectangle bounding boxes for textlines would never be a great option. So, we have developed an option which can try to find contours of those curvy textlines. You can set -cl (curved lines) to true to have this option. Be carefull that this increase the time, the tool needs to go through document.
|
||||
|
||||
* If you want to crop and save image regions inside document just provide a directory with this parameter, -si (save images).
|
||||
|
||||
* At the end this tool still needs to be optimized and developed. So if any problems occur or this tool performance does not meet your expectation, you can provide us your worthy feedback.
|
||||
* In cases where the documents include curved headers or curved lines it is obvious that rectangular bounding boxes for textlines will not be a great option. For this, we have developed an option which tries to find contours of the curvy textlines. You can set `-cl` (**c**urved **l**ines) to `true` to enable this option. Be advised that this will increase the time needed for the tool to process the document.
|
||||
|
||||
* If you want to crop and save image regions inside the document, just provide a directory with the parameter, `-si` (**s**ave **i**mages).
|
||||
|
||||
* This tool is actively being developed. If any problems occur or the performance does not meet your expectations, we welcome your feedback.
|
||||
|
|
|
|||
|
|
@ -1866,9 +1866,14 @@ class eynollah:
|
|||
|
||||
#all_box_coord.append(crop_coor)
|
||||
|
||||
mask_textline=np.zeros((textline_mask_tot_ea.shape))
|
||||
|
||||
mask_textline=cv2.fillPoly(mask_textline,pts=[contours_per_process[mv]],color=(1,1,1))
|
||||
|
||||
|
||||
|
||||
denoised=None
|
||||
all_text_region_raw=textline_mask_tot_ea[boxes_text[mv][1]:boxes_text[mv][1]+boxes_text[mv][3] , boxes_text[mv][0]:boxes_text[mv][0]+boxes_text[mv][2] ]
|
||||
all_text_region_raw=(textline_mask_tot_ea*mask_textline[:,:])[boxes_text[mv][1]:boxes_text[mv][1]+boxes_text[mv][3] , boxes_text[mv][0]:boxes_text[mv][0]+boxes_text[mv][2] ]
|
||||
all_text_region_raw=all_text_region_raw.astype(np.uint8)
|
||||
|
||||
img_int_p=all_text_region_raw[:,:]#self.all_text_region_raw[mv]
|
||||
|
|
@ -4232,106 +4237,106 @@ class eynollah:
|
|||
def find_num_col_deskew(self,regions_without_seperators,sigma_,multiplier=3.8 ):
|
||||
regions_without_seperators_0=regions_without_seperators[:,:].sum(axis=1)
|
||||
|
||||
meda_n_updown=regions_without_seperators_0[len(regions_without_seperators_0)::-1]
|
||||
##meda_n_updown=regions_without_seperators_0[len(regions_without_seperators_0)::-1]
|
||||
|
||||
first_nonzero=(next((i for i, x in enumerate(regions_without_seperators_0) if x), 0))
|
||||
last_nonzero=(next((i for i, x in enumerate(meda_n_updown) if x), 0))
|
||||
##first_nonzero=(next((i for i, x in enumerate(regions_without_seperators_0) if x), 0))
|
||||
##last_nonzero=(next((i for i, x in enumerate(meda_n_updown) if x), 0))
|
||||
|
||||
last_nonzero=len(regions_without_seperators_0)-last_nonzero
|
||||
##last_nonzero=len(regions_without_seperators_0)-last_nonzero
|
||||
|
||||
|
||||
y=regions_without_seperators_0#[first_nonzero:last_nonzero]
|
||||
|
||||
y_help=np.zeros(len(y)+20)
|
||||
##y_help=np.zeros(len(y)+20)
|
||||
|
||||
y_help[10:len(y)+10]=y
|
||||
##y_help[10:len(y)+10]=y
|
||||
|
||||
x=np.array( range(len(y)) )
|
||||
##x=np.array( range(len(y)) )
|
||||
|
||||
|
||||
|
||||
|
||||
zneg_rev=-y_help+np.max(y_help)
|
||||
##zneg_rev=-y_help+np.max(y_help)
|
||||
|
||||
zneg=np.zeros(len(zneg_rev)+20)
|
||||
##zneg=np.zeros(len(zneg_rev)+20)
|
||||
|
||||
zneg[10:len(zneg_rev)+10]=zneg_rev
|
||||
##zneg[10:len(zneg_rev)+10]=zneg_rev
|
||||
|
||||
z=gaussian_filter1d(y, sigma_)
|
||||
zneg= gaussian_filter1d(zneg, sigma_)
|
||||
###zneg= gaussian_filter1d(zneg, sigma_)
|
||||
|
||||
|
||||
peaks_neg, _ = find_peaks(zneg, height=0)
|
||||
peaks, _ = find_peaks(z, height=0)
|
||||
###peaks_neg, _ = find_peaks(zneg, height=0)
|
||||
###peaks, _ = find_peaks(z, height=0)
|
||||
|
||||
peaks_neg=peaks_neg-10-10
|
||||
###peaks_neg=peaks_neg-10-10
|
||||
|
||||
#print(np.std(z),'np.std(z)np.std(z)np.std(z)')
|
||||
####print(np.std(z),'np.std(z)np.std(z)np.std(z)')
|
||||
|
||||
##plt.plot(z)
|
||||
##plt.show()
|
||||
#####plt.plot(z)
|
||||
#####plt.show()
|
||||
|
||||
##plt.imshow(regions_without_seperators)
|
||||
##plt.show()
|
||||
"""
|
||||
last_nonzero=last_nonzero-0#100
|
||||
first_nonzero=first_nonzero+0#+100
|
||||
#####plt.imshow(regions_without_seperators)
|
||||
#####plt.show()
|
||||
###"""
|
||||
###last_nonzero=last_nonzero-0#100
|
||||
###first_nonzero=first_nonzero+0#+100
|
||||
|
||||
peaks_neg=peaks_neg[(peaks_neg>first_nonzero) & (peaks_neg<last_nonzero)]
|
||||
###peaks_neg=peaks_neg[(peaks_neg>first_nonzero) & (peaks_neg<last_nonzero)]
|
||||
|
||||
peaks=peaks[(peaks>.06*regions_without_seperators.shape[1]) & (peaks<0.94*regions_without_seperators.shape[1])]
|
||||
"""
|
||||
interest_pos=z[peaks]
|
||||
###peaks=peaks[(peaks>.06*regions_without_seperators.shape[1]) & (peaks<0.94*regions_without_seperators.shape[1])]
|
||||
###"""
|
||||
###interest_pos=z[peaks]
|
||||
|
||||
interest_pos=interest_pos[interest_pos>10]
|
||||
###interest_pos=interest_pos[interest_pos>10]
|
||||
|
||||
interest_neg=z[peaks_neg]
|
||||
###interest_neg=z[peaks_neg]
|
||||
|
||||
min_peaks_pos=np.mean(interest_pos)
|
||||
min_peaks_neg=0#np.min(interest_neg)
|
||||
###min_peaks_pos=np.mean(interest_pos)
|
||||
###min_peaks_neg=0#np.min(interest_neg)
|
||||
|
||||
dis_talaei=(min_peaks_pos-min_peaks_neg)/multiplier
|
||||
#print(interest_pos)
|
||||
grenze=min_peaks_pos-dis_talaei#np.mean(y[peaks_neg[0]:peaks_neg[len(peaks_neg)-1]])-np.std(y[peaks_neg[0]:peaks_neg[len(peaks_neg)-1]])/2.0
|
||||
###dis_talaei=(min_peaks_pos-min_peaks_neg)/multiplier
|
||||
####print(interest_pos)
|
||||
###grenze=min_peaks_pos-dis_talaei#np.mean(y[peaks_neg[0]:peaks_neg[len(peaks_neg)-1]])-np.std(y[peaks_neg[0]:peaks_neg[len(peaks_neg)-1]])/2.0
|
||||
|
||||
interest_neg_fin=interest_neg[(interest_neg<grenze)]
|
||||
peaks_neg_fin=peaks_neg[(interest_neg<grenze)]
|
||||
interest_neg_fin=interest_neg[(interest_neg<grenze)]
|
||||
###interest_neg_fin=interest_neg[(interest_neg<grenze)]
|
||||
###peaks_neg_fin=peaks_neg[(interest_neg<grenze)]
|
||||
###interest_neg_fin=interest_neg[(interest_neg<grenze)]
|
||||
|
||||
"""
|
||||
if interest_neg[0]<0.1:
|
||||
interest_neg=interest_neg[1:]
|
||||
if interest_neg[len(interest_neg)-1]<0.1:
|
||||
interest_neg=interest_neg[:len(interest_neg)-1]
|
||||
###"""
|
||||
###if interest_neg[0]<0.1:
|
||||
###interest_neg=interest_neg[1:]
|
||||
###if interest_neg[len(interest_neg)-1]<0.1:
|
||||
###interest_neg=interest_neg[:len(interest_neg)-1]
|
||||
|
||||
|
||||
|
||||
min_peaks_pos=np.min(interest_pos)
|
||||
min_peaks_neg=0#np.min(interest_neg)
|
||||
###min_peaks_pos=np.min(interest_pos)
|
||||
###min_peaks_neg=0#np.min(interest_neg)
|
||||
|
||||
|
||||
dis_talaei=(min_peaks_pos-min_peaks_neg)/multiplier
|
||||
grenze=min_peaks_pos-dis_talaei#np.mean(y[peaks_neg[0]:peaks_neg[len(peaks_neg)-1]])-np.std(y[peaks_neg[0]:peaks_neg[len(peaks_neg)-1]])/2.0
|
||||
"""
|
||||
#interest_neg_fin=interest_neg#[(interest_neg<grenze)]
|
||||
#peaks_neg_fin=peaks_neg#[(interest_neg<grenze)]
|
||||
#interest_neg_fin=interest_neg#[(interest_neg<grenze)]
|
||||
###dis_talaei=(min_peaks_pos-min_peaks_neg)/multiplier
|
||||
###grenze=min_peaks_pos-dis_talaei#np.mean(y[peaks_neg[0]:peaks_neg[len(peaks_neg)-1]])-np.std(y[peaks_neg[0]:peaks_neg[len(peaks_neg)-1]])/2.0
|
||||
###"""
|
||||
####interest_neg_fin=interest_neg#[(interest_neg<grenze)]
|
||||
####peaks_neg_fin=peaks_neg#[(interest_neg<grenze)]
|
||||
####interest_neg_fin=interest_neg#[(interest_neg<grenze)]
|
||||
|
||||
num_col=(len(interest_neg_fin))+1
|
||||
###num_col=(len(interest_neg_fin))+1
|
||||
|
||||
|
||||
p_l=0
|
||||
p_u=len(y)-1
|
||||
p_m=int(len(y)/2.)
|
||||
p_g_l=int(len(y)/3.)
|
||||
p_g_u=len(y)-int(len(y)/3.)
|
||||
###p_l=0
|
||||
###p_u=len(y)-1
|
||||
###p_m=int(len(y)/2.)
|
||||
###p_g_l=int(len(y)/3.)
|
||||
###p_g_u=len(y)-int(len(y)/3.)
|
||||
|
||||
|
||||
diff_peaks=np.abs( np.diff(peaks_neg_fin) )
|
||||
diff_peaks_annormal=diff_peaks[diff_peaks<30]
|
||||
###diff_peaks=np.abs( np.diff(peaks_neg_fin) )
|
||||
###diff_peaks_annormal=diff_peaks[diff_peaks<30]
|
||||
|
||||
#print(len(interest_neg_fin),np.mean(interest_neg_fin))
|
||||
return interest_neg_fin,np.std(z)
|
||||
return np.std(z)#interest_neg_fin,np.std(z)
|
||||
def return_deskew_slop(self,img_patch_org,sigma_des,main_page=False):
|
||||
|
||||
|
||||
|
|
@ -4358,13 +4363,27 @@ class eynollah:
|
|||
|
||||
img_int=np.zeros((img_patch_org.shape[0],img_patch_org.shape[1]))
|
||||
img_int[:,:]=img_patch_org[:,:]#img_patch_org[:,:,0]
|
||||
|
||||
|
||||
|
||||
max_shape=np.max(img_int.shape)
|
||||
img_resized=np.zeros((int( max_shape*(1.1) ) , int( max_shape*(1.1) ) ))
|
||||
|
||||
|
||||
onset_x=int((img_resized.shape[1]-img_int.shape[1])/2.)
|
||||
onset_y=int((img_resized.shape[0]-img_int.shape[0])/2.)
|
||||
|
||||
img_resized=np.zeros((int( img_int.shape[0]*(1.8) ) , int( img_int.shape[1]*(2.6) ) ))
|
||||
|
||||
#img_resized=np.zeros((int( img_int.shape[0]*(1.8) ) , int( img_int.shape[1]*(2.6) ) ))
|
||||
|
||||
|
||||
|
||||
img_resized[ int( img_int.shape[0]*(.4)):int( img_int.shape[0]*(.4))+img_int.shape[0] , int( img_int.shape[1]*(.8)):int( img_int.shape[1]*(.8))+img_int.shape[1] ]=img_int[:,:]
|
||||
#img_resized[ int( img_int.shape[0]*(.4)):int( img_int.shape[0]*(.4))+img_int.shape[0] , int( img_int.shape[1]*(.8)):int( img_int.shape[1]*(.8))+img_int.shape[1] ]=img_int[:,:]
|
||||
img_resized[ onset_y:onset_y+img_int.shape[0] , onset_x:onset_x+img_int.shape[1] ]=img_int[:,:]
|
||||
|
||||
#print(img_resized.shape,'img_resizedshape')
|
||||
#plt.imshow(img_resized)
|
||||
#plt.show()
|
||||
|
||||
if main_page and img_patch_org.shape[1]>img_patch_org.shape[0]:
|
||||
|
||||
|
|
@ -4372,12 +4391,12 @@ class eynollah:
|
|||
#plt.show()
|
||||
angels=np.array([-45, 0 , 45 , 90 , ])#np.linspace(-12,12,100)#np.array([0 , 45 , 90 , -45])
|
||||
|
||||
res=[]
|
||||
num_of_peaks=[]
|
||||
index_cor=[]
|
||||
#res=[]
|
||||
#num_of_peaks=[]
|
||||
#index_cor=[]
|
||||
var_res=[]
|
||||
|
||||
indexer=0
|
||||
#indexer=0
|
||||
for rot in angels:
|
||||
img_rot=self.rotate_image(img_resized,rot)
|
||||
#plt.imshow(img_rot)
|
||||
|
|
@ -4389,27 +4408,31 @@ class eynollah:
|
|||
#neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(var_spectrum,'var_spectrum')
|
||||
try:
|
||||
neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(rot,var_spectrum,'var_spectrum')
|
||||
res_me=np.mean(neg_peaks)
|
||||
if res_me==0:
|
||||
res_me=1000000000000000000000
|
||||
else:
|
||||
pass
|
||||
var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
##print(rot,var_spectrum,'var_spectrum')
|
||||
#res_me=np.mean(neg_peaks)
|
||||
#if res_me==0:
|
||||
#res_me=1000000000000000000000
|
||||
#else:
|
||||
#pass
|
||||
|
||||
res_num=len(neg_peaks)
|
||||
#res_num=len(neg_peaks)
|
||||
except:
|
||||
res_me=1000000000000000000000
|
||||
res_num=0
|
||||
#res_me=1000000000000000000000
|
||||
#res_num=0
|
||||
var_spectrum=0
|
||||
if self.isNaN(res_me):
|
||||
pass
|
||||
else:
|
||||
res.append( res_me )
|
||||
var_res.append(var_spectrum)
|
||||
num_of_peaks.append( res_num )
|
||||
index_cor.append(indexer)
|
||||
indexer=indexer+1
|
||||
#if self.isNaN(res_me):
|
||||
#pass
|
||||
#else:
|
||||
#res.append( res_me )
|
||||
#var_res.append(var_spectrum)
|
||||
#num_of_peaks.append( res_num )
|
||||
#index_cor.append(indexer)
|
||||
#indexer=indexer+1
|
||||
|
||||
var_res.append(var_spectrum)
|
||||
#index_cor.append(indexer)
|
||||
#indexer=indexer+1
|
||||
|
||||
|
||||
try:
|
||||
|
|
@ -4422,12 +4445,12 @@ class eynollah:
|
|||
|
||||
angels=np.linspace(ang_int-22.5,ang_int+22.5,100)
|
||||
|
||||
res=[]
|
||||
num_of_peaks=[]
|
||||
index_cor=[]
|
||||
#res=[]
|
||||
#num_of_peaks=[]
|
||||
#index_cor=[]
|
||||
var_res=[]
|
||||
|
||||
indexer=0
|
||||
|
||||
for rot in angels:
|
||||
img_rot=self.rotate_image(img_resized,rot)
|
||||
##plt.imshow(img_rot)
|
||||
|
|
@ -4435,27 +4458,13 @@ class eynollah:
|
|||
img_rot[img_rot!=0]=1
|
||||
#res_me=np.mean(self.find_num_col_deskew(img_rot,sigma_des,2.0 ))
|
||||
try:
|
||||
neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(indexer,'indexer')
|
||||
res_me=np.mean(neg_peaks)
|
||||
if res_me==0:
|
||||
res_me=1000000000000000000000
|
||||
else:
|
||||
pass
|
||||
|
||||
res_num=len(neg_peaks)
|
||||
var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
|
||||
except:
|
||||
res_me=1000000000000000000000
|
||||
res_num=0
|
||||
var_spectrum=0
|
||||
if self.isNaN(res_me):
|
||||
pass
|
||||
else:
|
||||
res.append( res_me )
|
||||
var_res.append(var_spectrum)
|
||||
num_of_peaks.append( res_num )
|
||||
index_cor.append(indexer)
|
||||
indexer=indexer+1
|
||||
|
||||
var_res.append(var_spectrum)
|
||||
|
||||
|
||||
|
||||
|
||||
|
|
@ -4472,12 +4481,9 @@ class eynollah:
|
|||
#plt.show()
|
||||
angels=np.linspace(-12,12,100)#np.array([0 , 45 , 90 , -45])
|
||||
|
||||
res=[]
|
||||
num_of_peaks=[]
|
||||
index_cor=[]
|
||||
|
||||
var_res=[]
|
||||
|
||||
indexer=0
|
||||
for rot in angels:
|
||||
img_rot=self.rotate_image(img_resized,rot)
|
||||
#plt.imshow(img_rot)
|
||||
|
|
@ -4489,30 +4495,16 @@ class eynollah:
|
|||
#neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(var_spectrum,'var_spectrum')
|
||||
try:
|
||||
neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(rot,var_spectrum,'var_spectrum')
|
||||
res_me=np.mean(neg_peaks)
|
||||
if res_me==0:
|
||||
res_me=1000000000000000000000
|
||||
else:
|
||||
pass
|
||||
|
||||
res_num=len(neg_peaks)
|
||||
var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
|
||||
except:
|
||||
res_me=1000000000000000000000
|
||||
res_num=0
|
||||
var_spectrum=0
|
||||
if self.isNaN(res_me):
|
||||
pass
|
||||
else:
|
||||
res.append( res_me )
|
||||
var_res.append(var_spectrum)
|
||||
num_of_peaks.append( res_num )
|
||||
index_cor.append(indexer)
|
||||
indexer=indexer+1
|
||||
|
||||
var_res.append(var_spectrum)
|
||||
|
||||
|
||||
if self.dir_of_all is not None:
|
||||
print('galdi?')
|
||||
#print('galdi?')
|
||||
plt.figure(figsize=(60,30))
|
||||
plt.rcParams['font.size']='50'
|
||||
plt.plot(angels,np.array(var_res),'-o',markersize=25,linewidth=4)
|
||||
|
|
@ -4537,12 +4529,8 @@ class eynollah:
|
|||
|
||||
angels=np.linspace(-90,-12,100)
|
||||
|
||||
res=[]
|
||||
num_of_peaks=[]
|
||||
index_cor=[]
|
||||
var_res=[]
|
||||
|
||||
indexer=0
|
||||
for rot in angels:
|
||||
img_rot=self.rotate_image(img_resized,rot)
|
||||
##plt.imshow(img_rot)
|
||||
|
|
@ -4550,27 +4538,11 @@ class eynollah:
|
|||
img_rot[img_rot!=0]=1
|
||||
#res_me=np.mean(self.find_num_col_deskew(img_rot,sigma_des,2.0 ))
|
||||
try:
|
||||
neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(indexer,'indexer')
|
||||
res_me=np.mean(neg_peaks)
|
||||
if res_me==0:
|
||||
res_me=1000000000000000000000
|
||||
else:
|
||||
pass
|
||||
|
||||
res_num=len(neg_peaks)
|
||||
var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
except:
|
||||
res_me=1000000000000000000000
|
||||
res_num=0
|
||||
var_spectrum=0
|
||||
if self.isNaN(res_me):
|
||||
pass
|
||||
else:
|
||||
res.append( res_me )
|
||||
var_res.append(var_spectrum)
|
||||
num_of_peaks.append( res_num )
|
||||
index_cor.append(indexer)
|
||||
indexer=indexer+1
|
||||
|
||||
var_res.append(var_spectrum)
|
||||
|
||||
|
||||
try:
|
||||
|
|
@ -4584,12 +4556,9 @@ class eynollah:
|
|||
|
||||
angels=np.linspace(90,12,100)
|
||||
|
||||
res=[]
|
||||
num_of_peaks=[]
|
||||
index_cor=[]
|
||||
|
||||
var_res=[]
|
||||
|
||||
indexer=0
|
||||
for rot in angels:
|
||||
img_rot=self.rotate_image(img_resized,rot)
|
||||
##plt.imshow(img_rot)
|
||||
|
|
@ -4597,27 +4566,12 @@ class eynollah:
|
|||
img_rot[img_rot!=0]=1
|
||||
#res_me=np.mean(self.find_num_col_deskew(img_rot,sigma_des,2.0 ))
|
||||
try:
|
||||
neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(indexer,'indexer')
|
||||
res_me=np.mean(neg_peaks)
|
||||
if res_me==0:
|
||||
res_me=1000000000000000000000
|
||||
else:
|
||||
pass
|
||||
|
||||
res_num=len(neg_peaks)
|
||||
except:
|
||||
res_me=1000000000000000000000
|
||||
res_num=0
|
||||
var_spectrum=0
|
||||
if self.isNaN(res_me):
|
||||
pass
|
||||
else:
|
||||
res.append( res_me )
|
||||
var_res.append(var_spectrum)
|
||||
num_of_peaks.append( res_num )
|
||||
index_cor.append(indexer)
|
||||
indexer=indexer+1
|
||||
|
||||
var_res.append(var_spectrum)
|
||||
|
||||
|
||||
try:
|
||||
|
|
@ -4631,9 +4585,6 @@ class eynollah:
|
|||
|
||||
angels=np.linspace(-25,25,60)
|
||||
|
||||
res=[]
|
||||
num_of_peaks=[]
|
||||
index_cor=[]
|
||||
var_res=[]
|
||||
|
||||
indexer=0
|
||||
|
|
@ -4648,27 +4599,11 @@ class eynollah:
|
|||
#neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(var_spectrum,'var_spectrum')
|
||||
try:
|
||||
neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(rot,var_spectrum,'var_spectrum')
|
||||
res_me=np.mean(neg_peaks)
|
||||
if res_me==0:
|
||||
res_me=1000000000000000000000
|
||||
else:
|
||||
pass
|
||||
|
||||
res_num=len(neg_peaks)
|
||||
var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
except:
|
||||
res_me=1000000000000000000000
|
||||
res_num=0
|
||||
var_spectrum=0
|
||||
if self.isNaN(res_me):
|
||||
pass
|
||||
else:
|
||||
res.append( res_me )
|
||||
var_res.append(var_spectrum)
|
||||
num_of_peaks.append( res_num )
|
||||
index_cor.append(indexer)
|
||||
indexer=indexer+1
|
||||
|
||||
var_res.append(var_spectrum)
|
||||
|
||||
|
||||
try:
|
||||
|
|
@ -4677,20 +4612,21 @@ class eynollah:
|
|||
ang_int=angels[np.argmax(var_res)]#angels_sorted[arg_final]#angels[arg_sort_early[arg_sort[arg_final]]]#angels[arg_fin]
|
||||
except:
|
||||
ang_int=0
|
||||
|
||||
#print(ang_int,'ang_int')
|
||||
|
||||
#plt.plot(var_res)
|
||||
#plt.show()
|
||||
|
||||
##plt.plot(mom3_res)
|
||||
##plt.show()
|
||||
#print(ang_int,'ang_int111')
|
||||
|
||||
early_slope_edge=22
|
||||
if abs(ang_int)>early_slope_edge and ang_int<0:
|
||||
|
||||
angels=np.linspace(-90,-25,60)
|
||||
|
||||
res=[]
|
||||
num_of_peaks=[]
|
||||
index_cor=[]
|
||||
var_res=[]
|
||||
|
||||
indexer=0
|
||||
for rot in angels:
|
||||
img_rot=self.rotate_image(img_resized,rot)
|
||||
##plt.imshow(img_rot)
|
||||
|
|
@ -4698,27 +4634,13 @@ class eynollah:
|
|||
img_rot[img_rot!=0]=1
|
||||
#res_me=np.mean(self.find_num_col_deskew(img_rot,sigma_des,2.0 ))
|
||||
try:
|
||||
neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(indexer,'indexer')
|
||||
res_me=np.mean(neg_peaks)
|
||||
if res_me==0:
|
||||
res_me=1000000000000000000000
|
||||
else:
|
||||
pass
|
||||
|
||||
res_num=len(neg_peaks)
|
||||
var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
|
||||
except:
|
||||
res_me=1000000000000000000000
|
||||
res_num=0
|
||||
var_spectrum=0
|
||||
if self.isNaN(res_me):
|
||||
pass
|
||||
else:
|
||||
res.append( res_me )
|
||||
var_res.append(var_spectrum)
|
||||
num_of_peaks.append( res_num )
|
||||
index_cor.append(indexer)
|
||||
indexer=indexer+1
|
||||
|
||||
var_res.append(var_spectrum)
|
||||
|
||||
|
||||
|
||||
try:
|
||||
|
|
@ -4732,9 +4654,6 @@ class eynollah:
|
|||
|
||||
angels=np.linspace(90,25,60)
|
||||
|
||||
res=[]
|
||||
num_of_peaks=[]
|
||||
index_cor=[]
|
||||
var_res=[]
|
||||
|
||||
indexer=0
|
||||
|
|
@ -4745,27 +4664,13 @@ class eynollah:
|
|||
img_rot[img_rot!=0]=1
|
||||
#res_me=np.mean(self.find_num_col_deskew(img_rot,sigma_des,2.0 ))
|
||||
try:
|
||||
neg_peaks,var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
var_spectrum=self.find_num_col_deskew(img_rot,sigma_des,20.3 )
|
||||
#print(indexer,'indexer')
|
||||
res_me=np.mean(neg_peaks)
|
||||
if res_me==0:
|
||||
res_me=1000000000000000000000
|
||||
else:
|
||||
pass
|
||||
|
||||
res_num=len(neg_peaks)
|
||||
except:
|
||||
res_me=1000000000000000000000
|
||||
res_num=0
|
||||
var_spectrum=0
|
||||
if self.isNaN(res_me):
|
||||
pass
|
||||
else:
|
||||
res.append( res_me )
|
||||
var_res.append(var_spectrum)
|
||||
num_of_peaks.append( res_num )
|
||||
index_cor.append(indexer)
|
||||
indexer=indexer+1
|
||||
|
||||
var_res.append(var_spectrum)
|
||||
|
||||
|
||||
|
||||
try:
|
||||
|
|
@ -10443,7 +10348,9 @@ class eynollah:
|
|||
if gaussian_filter:
|
||||
img= cv2.GaussianBlur(img,(5,5),0)
|
||||
img = img.astype(np.uint16)
|
||||
prediction_regions_org2=self.do_prediction(patches,img,model_region)
|
||||
|
||||
marginal_patch=0.2
|
||||
prediction_regions_org2=self.do_prediction(patches,img,model_region,marginal_patch)
|
||||
|
||||
prediction_regions_org2=self.resize_image(prediction_regions_org2, img_height_h, img_width_h )
|
||||
|
||||
|
|
@ -10825,6 +10732,8 @@ class eynollah:
|
|||
|
||||
text_regions[(mask_marginals_rotated[:,:]!=1) & (text_regions[:,:]==1)]=4
|
||||
|
||||
#plt.imshow(text_regions)
|
||||
#plt.show()
|
||||
|
||||
pixel_img=4
|
||||
min_area_text=0.00001
|
||||
|
|
@ -10843,8 +10752,8 @@ class eynollah:
|
|||
|
||||
x_width_mar=abs(x_min_text_only[i]-x_max_text_only[i])
|
||||
y_height_mar=abs(y_min_text_only[i]-y_max_text_only[i])
|
||||
#print(x_width_mar,y_height_mar,'y_height_mar')
|
||||
if x_width_mar>16 and y_height_mar/x_width_mar<10:
|
||||
#print(x_width_mar,y_height_mar,y_height_mar/x_width_mar,'y_height_mar')
|
||||
if x_width_mar>16 and y_height_mar/x_width_mar<18:
|
||||
marginlas_should_be_main_text.append(polygons_of_marginals[i])
|
||||
if x_min_text_only[i]<(mid_point-one_third_left):
|
||||
x_min_marginals_left_new=x_min_text_only[i]
|
||||
|
|
@ -11115,8 +11024,8 @@ class eynollah:
|
|||
textlines_con_changed.append(textlines_big_org_form)
|
||||
return textlines_con_changed
|
||||
def check_any_text_region_in_model_one_is_main_or_header(self,regions_model_1,regions_model_full,contours_only_text_parent,all_box_coord,all_found_texline_polygons,slopes,contours_only_text_parent_d_ordered):
|
||||
text_only=(regions_model_1[:,:]==1)*1
|
||||
contours_only_text,hir_on_text=self.return_contours_of_image(text_only)
|
||||
#text_only=(regions_model_1[:,:]==1)*1
|
||||
#contours_only_text,hir_on_text=self.return_contours_of_image(text_only)
|
||||
|
||||
"""
|
||||
contours_only_text_parent=self.return_parent_contours( contours_only_text,hir_on_text)
|
||||
|
|
@ -12290,7 +12199,7 @@ class eynollah:
|
|||
num_col=None
|
||||
peaks_neg_fin=[]
|
||||
|
||||
print(num_col,'num_colnum_col')
|
||||
#print(num_col,'num_colnum_col')
|
||||
if num_col is None:
|
||||
txt_con_org=[]
|
||||
order_text_new=[]
|
||||
|
|
@ -12316,7 +12225,7 @@ class eynollah:
|
|||
K.clear_session()
|
||||
gc.collect()
|
||||
|
||||
print(np.unique(textline_mask_tot_ea[:,:]),'textline')
|
||||
#print(np.unique(textline_mask_tot_ea[:,:]),'textline')
|
||||
|
||||
if self.dir_of_all is not None:
|
||||
|
||||
|
|
@ -12771,7 +12680,7 @@ class eynollah:
|
|||
|
||||
|
||||
slopes_marginals,all_found_texline_polygons_marginals,boxes_marginals,_,polygons_of_marginals,all_box_coord_marginals,index_by_text_par_con_marginal=self.get_slopes_and_deskew_new(polygons_of_marginals,polygons_of_marginals,textline_mask_tot_ea,image_page_rotated,boxes_marginals,slope_deskew)
|
||||
|
||||
|
||||
|
||||
if self.curved_line=='true' or self.curved_line=='True' or self.curved_line=='TRUE' :
|
||||
scale_param=1
|
||||
|
|
@ -12911,9 +12820,15 @@ class eynollah:
|
|||
boxes=self.return_boxes_of_images_by_order_of_reading_new(spliter_y_new,regions_without_seperators,matrix_of_lines_ch)
|
||||
else:
|
||||
boxes_d=self.return_boxes_of_images_by_order_of_reading_new(spliter_y_new_d,regions_without_seperators_d,matrix_of_lines_ch_d)
|
||||
|
||||
#print(boxes_d,len(boxes_d),'boxes_d')
|
||||
#for mv in range(len(boxes_d)):
|
||||
|
||||
#img_box=image_page[boxes_d[mv][1]:boxes_d[mv][1]+boxes_d[mv][3] , boxes_d[mv][0]:boxes_d[mv][0]+boxes_d[mv][2],: ]
|
||||
#plt.imshow(img_box)
|
||||
#plt.show()
|
||||
|
||||
|
||||
|
||||
#print(slopes)
|
||||
if self.dir_of_cropped_images is not None:
|
||||
self.write_images_into_directory(polygons_of_images,self.dir_of_cropped_images,image_page)
|
||||
|
|
|
|||
Loading…
Add table
Add a link
Reference in a new issue