mirror of
https://github.com/qurator-spk/eynollah.git
synced 2026-03-06 15:21:58 +01:00
Compare commits
No commits in common. "main" and "v0.4.0" have entirely different histories.
94 changed files with 5614 additions and 19767 deletions
2
.github/workflows/build-docker.yml
vendored
2
.github/workflows/build-docker.yml
vendored
|
|
@ -2,7 +2,7 @@ name: CD
|
|||
|
||||
on:
|
||||
push:
|
||||
branches: [ "main" ]
|
||||
branches: [ "master" ]
|
||||
workflow_dispatch: # run manually
|
||||
|
||||
jobs:
|
||||
|
|
|
|||
24
.github/workflows/pypi.yml
vendored
24
.github/workflows/pypi.yml
vendored
|
|
@ -1,24 +0,0 @@
|
|||
name: PyPI CD
|
||||
|
||||
on:
|
||||
release:
|
||||
types: [published]
|
||||
workflow_dispatch:
|
||||
|
||||
jobs:
|
||||
pypi-publish:
|
||||
name: upload release to PyPI
|
||||
runs-on: ubuntu-latest
|
||||
permissions:
|
||||
# IMPORTANT: this permission is mandatory for Trusted Publishing
|
||||
id-token: write
|
||||
steps:
|
||||
- uses: actions/checkout@v4
|
||||
- name: Set up Python
|
||||
uses: actions/setup-python@v5
|
||||
- name: Build package
|
||||
run: make build
|
||||
- name: Publish package distributions to PyPI
|
||||
uses: pypa/gh-action-pypi-publish@release/v1
|
||||
with:
|
||||
verbose: true
|
||||
48
.github/workflows/test-eynollah.yml
vendored
48
.github/workflows/test-eynollah.yml
vendored
|
|
@ -24,59 +24,36 @@ jobs:
|
|||
sudo rm -rf "$AGENT_TOOLSDIRECTORY"
|
||||
df -h
|
||||
- uses: actions/checkout@v4
|
||||
|
||||
# - name: Lint with ruff
|
||||
# uses: astral-sh/ruff-action@v3
|
||||
# with:
|
||||
# src: "./src"
|
||||
|
||||
- name: Try to restore models_eynollah
|
||||
uses: actions/cache/restore@v4
|
||||
id: all_model_cache
|
||||
- uses: actions/cache@v4
|
||||
id: seg_model_cache
|
||||
with:
|
||||
path: models_eynollah
|
||||
key: models_eynollah-${{ hashFiles('src/eynollah/model_zoo/default_specs.py') }}
|
||||
|
||||
key: ${{ runner.os }}-models
|
||||
- uses: actions/cache@v4
|
||||
id: bin_model_cache
|
||||
with:
|
||||
path: default-2021-03-09
|
||||
key: ${{ runner.os }}-modelbin
|
||||
- name: Download models
|
||||
if: steps.all_model_cache.outputs.cache-hit != 'true'
|
||||
run: |
|
||||
make models
|
||||
ls -la models_eynollah
|
||||
|
||||
- uses: actions/cache/save@v4
|
||||
if: steps.all_model_cache.outputs.cache-hit != 'true'
|
||||
with:
|
||||
path: models_eynollah
|
||||
key: models_eynollah-${{ hashFiles('src/eynollah/model_zoo/default_specs.py') }}
|
||||
|
||||
if: steps.seg_model_cache.outputs.cache-hit != 'true' || steps.bin_model_cache.outputs.cache-hit != 'true'
|
||||
run: make models
|
||||
- name: Set up Python ${{ matrix.python-version }}
|
||||
uses: actions/setup-python@v5
|
||||
with:
|
||||
python-version: ${{ matrix.python-version }}
|
||||
|
||||
# - uses: actions/cache@v4
|
||||
# with:
|
||||
# path: |
|
||||
# path/to/dependencies
|
||||
# some/other/dependencies
|
||||
# key: ${{ runner.os }}-${{ hashFiles('**/lockfiles') }}
|
||||
|
||||
- name: Install dependencies
|
||||
run: |
|
||||
python -m pip install --upgrade pip
|
||||
make install-dev EXTRAS=OCR,plotting
|
||||
make deps-test EXTRAS=OCR,plotting
|
||||
|
||||
make deps-test
|
||||
- name: Test with pytest
|
||||
run: make coverage PYTEST_ARGS="-vv --junitxml=pytest.xml"
|
||||
|
||||
- name: Get coverage results
|
||||
run: |
|
||||
coverage report --format=markdown >> $GITHUB_STEP_SUMMARY
|
||||
coverage html
|
||||
coverage json
|
||||
coverage xml
|
||||
|
||||
- name: Store coverage results
|
||||
uses: actions/upload-artifact@v4
|
||||
with:
|
||||
|
|
@ -86,15 +63,12 @@ jobs:
|
|||
pytest.xml
|
||||
coverage.xml
|
||||
coverage.json
|
||||
|
||||
- name: Upload coverage results
|
||||
uses: codecov/codecov-action@v4
|
||||
with:
|
||||
files: coverage.xml
|
||||
fail_ci_if_error: false
|
||||
|
||||
- name: Test standalone CLI
|
||||
run: make smoke-test
|
||||
|
||||
- name: Test OCR-D CLI
|
||||
run: make ocrd-test
|
||||
|
|
|
|||
7
.gitignore
vendored
7
.gitignore
vendored
|
|
@ -2,13 +2,6 @@
|
|||
__pycache__
|
||||
sbb_newspapers_org_image/pylint.log
|
||||
models_eynollah*
|
||||
models_ocr*
|
||||
models_layout*
|
||||
default-2021-03-09
|
||||
output.html
|
||||
/build
|
||||
/dist
|
||||
*.tif
|
||||
*.sw?
|
||||
TAGS
|
||||
uv.lock
|
||||
|
|
|
|||
143
CHANGELOG.md
143
CHANGELOG.md
|
|
@ -5,147 +5,12 @@ Versioned according to [Semantic Versioning](http://semver.org/).
|
|||
|
||||
## Unreleased
|
||||
|
||||
Added:
|
||||
|
||||
* "Model zoo", central place to describe and load models, #207
|
||||
* Training code for the CNN/RNN OCR model
|
||||
|
||||
Changed:
|
||||
|
||||
* Lint training code, #204
|
||||
* Update documentation: README, pyproject.toml metadata, guides in `docs/`, #209
|
||||
|
||||
|
||||
## [0.6.0] - 2025-10-17
|
||||
|
||||
Added:
|
||||
|
||||
* `eynollah-training` CLI and docs for training the models, #187, #193, https://github.com/qurator-spk/sbb_pixelwise_segmentation/tree/unifying-training-models
|
||||
|
||||
Fixed:
|
||||
|
||||
* `join_polygons` always returning Polygon, not MultiPolygon, #203
|
||||
|
||||
## [0.6.0rc2] - 2025-10-14
|
||||
|
||||
Fixed:
|
||||
|
||||
* Prevent OOM GPU error by avoiding loading the `region_fl` model, #199
|
||||
* XML output: encoding should be `utf-8`, not `utf8`, #196, #197
|
||||
|
||||
## [0.6.0rc1] - 2025-10-10
|
||||
|
||||
Fixed:
|
||||
|
||||
* continue processing when no columns detected but text regions exist
|
||||
* convert marginalia to main text if no main text is present
|
||||
* reset deskewing angle to 0° when text covers <30% image area and detected angle >45°
|
||||
* :fire: polygons: avoid invalid paths (use `Polygon.buffer()` instead of dilation etc.)
|
||||
* `return_boxes_of_images_by_order_of_reading_new`: avoid Numpy.dtype mismatch, simplify
|
||||
* `return_boxes_of_images_by_order_of_reading_new`: log any exceptions instead of ignoring
|
||||
* `filter_contours_without_textline_inside`: avoid removing from duplicate lists twice
|
||||
* `get_marginals`: exit early if no peaks found to avoid spurious overlap mask
|
||||
* `get_smallest_skew`: after shifting search range of rotation angle, use overall best result
|
||||
* Dockerfile: fix CUDA installation (cuDNN contested between Torch and TF due to extra OCR)
|
||||
* OCR: re-instate missing methods and fix `utils_ocr` function calls
|
||||
* mbreorder/enhancement CLIs: missing imports
|
||||
* :fire: writer: `SeparatorRegion` needs `SeparatorRegionType` (not `ImageRegionType`), f458e3e
|
||||
* tests: switch from `pytest-subtests` to `parametrize` so we can use `pytest-isolate`
|
||||
(so CUDA memory gets freed between tests if running on GPU)
|
||||
|
||||
Added:
|
||||
* :fire: `layout` CLI: new option `--model_version` to override default choices
|
||||
* test coverage for OCR options in `layout`
|
||||
* test coverage for table detection in `layout`
|
||||
* CI linting with ruff
|
||||
|
||||
Changed:
|
||||
|
||||
* polygons: slightly widen for regions and lines, increase for separators
|
||||
* various refactorings, some code style and identifier improvements
|
||||
* deskewing/multiprocessing: switch back to ProcessPoolExecutor (faster),
|
||||
but use shared memory if necessary, and switch back from `loky` to stdlib,
|
||||
and shutdown in `del()` instead of `atexit`
|
||||
* :fire: OCR: switch CNN-RNN model to `20250930` version compatible with TF 2.12 on CPU, too
|
||||
* OCR: allow running `-tr` without `-fl`, too
|
||||
* :fire: writer: use `@type='heading'` instead of `'header'` for headings
|
||||
* :fire: performance gains via refactoring (simplification, less copy-code, vectorization,
|
||||
avoiding unused calculations, avoiding unnecessary 3-channel image operations)
|
||||
* :fire: heuristic reading order detection: many improvements
|
||||
- contour vs splitter box matching:
|
||||
* contour must be contained in box exactly instead of heuristics
|
||||
* make fallback center matching, center must be contained in box
|
||||
- original vs deskewed contour matching:
|
||||
* same min-area filter on both sides
|
||||
* similar area score in addition to center proximity
|
||||
* avoid duplicate and missing mappings by allowing N:M
|
||||
matches and splitting+joining where necessary
|
||||
* CI: update+improve model caching
|
||||
|
||||
|
||||
## [0.5.0] - 2025-09-26
|
||||
|
||||
Fixed:
|
||||
|
||||
* restoring the contour in the original image caused an error due to an empty tuple, #154
|
||||
* removed NumPy warnings calculating sigma, mean, (fixed issue #158)
|
||||
* fixed bug in `separate_lines.py`, #124
|
||||
* Drop capitals are now handled separately from their corresponding textline
|
||||
* Marginals are now divided into left and right. Their reading order is written first for left marginals, then for right marginals, and within each side from top to bottom
|
||||
* Added a new page extraction model. Instead of bounding boxes, it outputs page contours in the XML file, improving results for skewed pages
|
||||
* Improved reading order for cases where a textline is segmented into multiple smaller textlines
|
||||
|
||||
Changed
|
||||
|
||||
* CLIs: read only allowed filename suffixes (image or XML) with `--dir_in`
|
||||
* CLIs: make all output option required, and `-i` / `-di` required but mutually exclusive
|
||||
* ocr CLI: drop redundant `-brb` in favour of just `-dib`
|
||||
* APIs: move all input/output path options from class (kwarg and attribute) ro `run` kwarg
|
||||
* layout textlines: polygonal also without `-cl`
|
||||
|
||||
Added:
|
||||
|
||||
* `eynollah machine-based-reading-order` CLI to run reading order detection, #175
|
||||
* `eynollah enhancement` CLI to run image enhancement, #175
|
||||
* Improved models for page extraction and reading order detection, #175
|
||||
* For the lightweight version (layout and textline detection), thresholds are now assigned to the artificial class. Users can apply these thresholds to improve detection of isolated textlines and regions. To counteract the drawback of thresholding, the skeleton of the artificial class is used to keep lines as thin as possible (resolved issues #163 and #161)
|
||||
* Added and integrated a trained CNN-RNN OCR models
|
||||
* Added and integrated a trained TrOCR model
|
||||
* Improved OCR detection to support vertical and curved textlines
|
||||
* Introduced a new machine-based reading order model with rotation augmentation
|
||||
* Optimized reading order speed by clustering text regions that belong to the same block, maintaining top-to-bottom order
|
||||
* Implemented text merging across textlines based on hyphenation when a line ends with a hyphen
|
||||
* Integrated image enhancement as a separate use case
|
||||
* Added reading order functionality on the layout level as a separate use case
|
||||
* CNN-RNN OCR models provide confidence scores for predictions
|
||||
* Added OCR visualization: predicted OCR can be overlaid on an image of the same size as the input
|
||||
* Introduced a threshold value for CNN-RNN OCR models, allowing users to filter out low-confidence textline predictions
|
||||
* For OCR, users can specify a single model by name instead of always using the default model
|
||||
* Under the OCR use case, if Ground Truth XMLs and images are available, textline image and corresponding text extraction can now be performed
|
||||
|
||||
Merged PRs:
|
||||
|
||||
* better machine based reading order + layout and textline + ocr by @vahidrezanezhad in https://github.com/qurator-spk/eynollah/pull/175
|
||||
* CI: pypi by @kba in https://github.com/qurator-spk/eynollah/pull/154
|
||||
* CI: Use most recent actions/setup-python@v5 by @kba in https://github.com/qurator-spk/eynollah/pull/157
|
||||
* update docker by @bertsky in https://github.com/qurator-spk/eynollah/pull/159
|
||||
* Ocrd fixes by @kba in https://github.com/qurator-spk/eynollah/pull/167
|
||||
* Updating readme for eynollah use cases cli by @kba in https://github.com/qurator-spk/eynollah/pull/166
|
||||
* OCR-D processor: expose reading_order_machine_based by @bertsky in https://github.com/qurator-spk/eynollah/pull/171
|
||||
* prepare release v0.5.0: fix logging by @bertsky in https://github.com/qurator-spk/eynollah/pull/180
|
||||
* mb_ro_on_layout: remove copy-pasta code not actually used by @kba in https://github.com/qurator-spk/eynollah/pull/181
|
||||
* prepare release v0.5.0: improve CLI docstring, refactor I/O path options from class to run kwargs, increase test coverage @bertsky in #182
|
||||
* prepare release v0.5.0: fix for OCR doit subtest by @bertsky in https://github.com/qurator-spk/eynollah/pull/183
|
||||
* Prepare release v0.5.0 by @kba in https://github.com/qurator-spk/eynollah/pull/178
|
||||
* updating eynollah README, how to use it for use cases by @vahidrezanezhad in https://github.com/qurator-spk/eynollah/pull/156
|
||||
* add feedback to command line interface by @michalbubula in https://github.com/qurator-spk/eynollah/pull/170
|
||||
|
||||
## [0.4.0] - 2025-04-07
|
||||
|
||||
Fixed:
|
||||
|
||||
* allow empty imports for optional dependencies
|
||||
* avoid Numpy warnings (empty slices etc.)
|
||||
* avoid Numpy warnings (empty slices etc)
|
||||
* remove deprecated Numpy types
|
||||
* binarization CLI: make `dir_in` usable again
|
||||
|
||||
|
|
@ -318,12 +183,6 @@ Fixed:
|
|||
Initial release
|
||||
|
||||
<!-- link-labels -->
|
||||
[0.7.0]: ../../compare/v0.7.0...v0.6.0
|
||||
[0.6.0]: ../../compare/v0.6.0...v0.6.0rc2
|
||||
[0.6.0rc2]: ../../compare/v0.6.0rc2...v0.6.0rc1
|
||||
[0.6.0rc1]: ../../compare/v0.6.0rc1...v0.5.0
|
||||
[0.5.0]: ../../compare/v0.5.0...v0.4.0
|
||||
[0.4.0]: ../../compare/v0.4.0...v0.3.1
|
||||
[0.3.1]: ../../compare/v0.3.1...v0.3.0
|
||||
[0.3.0]: ../../compare/v0.3.0...v0.2.0
|
||||
[0.2.0]: ../../compare/v0.2.0...v0.1.0
|
||||
|
|
|
|||
|
|
@ -36,12 +36,8 @@ COPY . .
|
|||
COPY ocrd-tool.json .
|
||||
# prepackage ocrd-tool.json as ocrd-all-tool.json
|
||||
RUN ocrd ocrd-tool ocrd-tool.json dump-tools > $(dirname $(ocrd bashlib filename))/ocrd-all-tool.json
|
||||
# prepackage ocrd-all-module-dir.json
|
||||
RUN ocrd ocrd-tool ocrd-tool.json dump-module-dirs > $(dirname $(ocrd bashlib filename))/ocrd-all-module-dir.json
|
||||
# install everything and reduce image size
|
||||
RUN make install EXTRAS=OCR && rm -rf /build/eynollah
|
||||
# fixup for broken cuDNN installation (Torch pulls in 8.5.0, which is incompatible with Tensorflow)
|
||||
RUN pip install nvidia-cudnn-cu11==8.6.0.163
|
||||
# smoke test
|
||||
RUN eynollah --help
|
||||
|
||||
|
|
|
|||
77
Makefile
77
Makefile
|
|
@ -3,22 +3,18 @@ PIP ?= pip3
|
|||
EXTRAS ?=
|
||||
|
||||
# DOCKER_BASE_IMAGE = artefakt.dev.sbb.berlin:5000/sbb/ocrd_core:v2.68.0
|
||||
DOCKER_BASE_IMAGE ?= docker.io/ocrd/core-cuda-tf2:latest
|
||||
DOCKER_TAG ?= ocrd/eynollah
|
||||
DOCKER ?= docker
|
||||
WGET = wget -O
|
||||
DOCKER_BASE_IMAGE = docker.io/ocrd/core-cuda-tf2:v3.3.0
|
||||
DOCKER_TAG = ocrd/eynollah
|
||||
|
||||
#SEG_MODEL := https://qurator-data.de/eynollah/2021-04-25/models_eynollah.tar.gz
|
||||
#SEG_MODEL := https://qurator-data.de/eynollah/2022-04-05/models_eynollah_renamed.tar.gz
|
||||
# SEG_MODEL := https://qurator-data.de/eynollah/2022-04-05/models_eynollah.tar.gz
|
||||
SEG_MODEL := https://qurator-data.de/eynollah/2022-04-05/models_eynollah.tar.gz
|
||||
#SEG_MODEL := https://github.com/qurator-spk/eynollah/releases/download/v0.3.0/models_eynollah.tar.gz
|
||||
#SEG_MODEL := https://github.com/qurator-spk/eynollah/releases/download/v0.3.1/models_eynollah.tar.gz
|
||||
#SEG_MODEL := https://zenodo.org/records/17194824/files/models_layout_v0_5_0.tar.gz?download=1
|
||||
EYNOLLAH_MODELS_URL := https://zenodo.org/records/17580627/files/models_all_v0_7_0.zip
|
||||
EYNOLLAH_MODELS_ZIP = $(notdir $(EYNOLLAH_MODELS_URL))
|
||||
EYNOLLAH_MODELS_DIR = $(EYNOLLAH_MODELS_ZIP:%.zip=%)
|
||||
|
||||
PYTEST_ARGS ?= -vv --isolate
|
||||
BIN_MODEL := https://github.com/qurator-spk/sbb_binarization/releases/download/v0.0.11/saved_model_2021_03_09.zip
|
||||
|
||||
PYTEST_ARGS ?= -vv
|
||||
|
||||
# BEGIN-EVAL makefile-parser --make-help Makefile
|
||||
|
||||
|
|
@ -31,8 +27,7 @@ help:
|
|||
@echo " install Install package with pip"
|
||||
@echo " install-dev Install editable with pip"
|
||||
@echo " deps-test Install test dependencies with pip"
|
||||
@echo " models Download and extract models to $(CURDIR):"
|
||||
@echo " $(EYNOLLAH_MODELS_DIR)"
|
||||
@echo " models Download and extract models to $(CURDIR)/models_eynollah"
|
||||
@echo " smoke-test Run simple CLI check"
|
||||
@echo " ocrd-test Run OCR-D CLI check"
|
||||
@echo " test Run unit tests"
|
||||
|
|
@ -41,22 +36,29 @@ help:
|
|||
@echo " EXTRAS comma-separated list of features (like 'OCR,plotting') for 'install' [$(EXTRAS)]"
|
||||
@echo " DOCKER_TAG Docker image tag for 'docker' [$(DOCKER_TAG)]"
|
||||
@echo " PYTEST_ARGS pytest args for 'test' (Set to '-s' to see log output during test execution, '-vv' to see individual tests. [$(PYTEST_ARGS)]"
|
||||
@echo " ALL_MODELS URL of archive of all models [$(ALL_MODELS)]"
|
||||
@echo " SEG_MODEL URL of 'models' archive to download for segmentation 'test' [$(SEG_MODEL)]"
|
||||
@echo " BIN_MODEL URL of 'models' archive to download for binarization 'test' [$(BIN_MODEL)]"
|
||||
@echo ""
|
||||
|
||||
# END-EVAL
|
||||
|
||||
# Download and extract models to $(PWD)/models_layout_v0_6_0
|
||||
models: $(EYNOLLAH_MODELS_DIR)
|
||||
|
||||
# do not download these files if we already have the directories
|
||||
.INTERMEDIATE: $(EYNOLLAH_MODELS_ZIP)
|
||||
# Download and extract models to $(PWD)/models_eynollah
|
||||
models: models_eynollah default-2021-03-09
|
||||
|
||||
$(EYNOLLAH_MODELS_ZIP):
|
||||
$(WGET) $@ $(EYNOLLAH_MODELS_URL)
|
||||
models_eynollah: models_eynollah.tar.gz
|
||||
tar zxf models_eynollah.tar.gz
|
||||
|
||||
$(EYNOLLAH_MODELS_DIR): $(EYNOLLAH_MODELS_ZIP)
|
||||
unzip $<
|
||||
models_eynollah.tar.gz:
|
||||
wget $(SEG_MODEL)
|
||||
|
||||
default-2021-03-09: $(notdir $(BIN_MODEL))
|
||||
unzip $(notdir $(BIN_MODEL))
|
||||
mkdir $@
|
||||
mv $(basename $(notdir $(BIN_MODEL))) $@
|
||||
|
||||
$(notdir $(BIN_MODEL)):
|
||||
wget $(BIN_MODEL)
|
||||
|
||||
build:
|
||||
$(PIP) install build
|
||||
|
|
@ -70,48 +72,41 @@ install:
|
|||
install-dev:
|
||||
$(PIP) install -e .$(and $(EXTRAS),[$(EXTRAS)])
|
||||
|
||||
deps-test:
|
||||
deps-test: models_eynollah
|
||||
$(PIP) install -r requirements-test.txt
|
||||
|
||||
smoke-test: TMPDIR != mktemp -d
|
||||
smoke-test: tests/resources/2files/kant_aufklaerung_1784_0020.tif
|
||||
smoke-test: tests/resources/kant_aufklaerung_1784_0020.tif
|
||||
# layout analysis:
|
||||
eynollah -m $(CURDIR) layout -i $< -o $(TMPDIR)
|
||||
eynollah layout -i $< -o $(TMPDIR) -m $(CURDIR)/models_eynollah
|
||||
fgrep -q http://schema.primaresearch.org/PAGE/gts/pagecontent/2019-07-15 $(TMPDIR)/$(basename $(<F)).xml
|
||||
fgrep -c -e TextRegion -e ImageRegion -e SeparatorRegion $(TMPDIR)/$(basename $(<F)).xml
|
||||
# layout, directory mode (skip one, add one):
|
||||
eynollah -m $(CURDIR) layout -di $(<D) -o $(TMPDIR)
|
||||
# directory mode (skip one, add one):
|
||||
eynollah layout -di $(<D) -o $(TMPDIR) -m $(CURDIR)/models_eynollah
|
||||
test -s $(TMPDIR)/euler_rechenkunst01_1738_0025.xml
|
||||
# mbreorder, directory mode (overwrite):
|
||||
eynollah -m $(CURDIR) machine-based-reading-order -di $(<D) -o $(TMPDIR)
|
||||
fgrep -q http://schema.primaresearch.org/PAGE/gts/pagecontent/2019-07-15 $(TMPDIR)/$(basename $(<F)).xml
|
||||
fgrep -c -e RegionRefIndexed $(TMPDIR)/$(basename $(<F)).xml
|
||||
# binarize:
|
||||
eynollah -m $(CURDIR) binarization -i $< -o $(TMPDIR)/$(<F)
|
||||
test -s $(TMPDIR)/$(<F)
|
||||
@set -x; test "$$(identify -format '%w %h' $<)" = "$$(identify -format '%w %h' $(TMPDIR)/$(<F))"
|
||||
# enhance:
|
||||
eynollah -m $(CURDIR) enhancement -sos -i $< -o $(TMPDIR) -O
|
||||
eynollah binarization -m $(CURDIR)/default-2021-03-09 $< $(TMPDIR)/$(<F)
|
||||
test -s $(TMPDIR)/$(<F)
|
||||
@set -x; test "$$(identify -format '%w %h' $<)" = "$$(identify -format '%w %h' $(TMPDIR)/$(<F))"
|
||||
$(RM) -r $(TMPDIR)
|
||||
|
||||
ocrd-test: export OCRD_MISSING_OUTPUT := ABORT
|
||||
ocrd-test: TMPDIR != mktemp -d
|
||||
ocrd-test: tests/resources/2files/kant_aufklaerung_1784_0020.tif
|
||||
ocrd-test: tests/resources/kant_aufklaerung_1784_0020.tif
|
||||
cp $< $(TMPDIR)
|
||||
ocrd workspace -d $(TMPDIR) init
|
||||
ocrd workspace -d $(TMPDIR) add -G OCR-D-IMG -g PHYS_0020 -i OCR-D-IMG_0020 $(<F)
|
||||
ocrd-eynollah-segment -w $(TMPDIR) -I OCR-D-IMG -O OCR-D-SEG -P models $(CURDIR)
|
||||
ocrd-eynollah-segment -w $(TMPDIR) -I OCR-D-IMG -O OCR-D-SEG -P models $(CURDIR)/models_eynollah
|
||||
result=$$(ocrd workspace -d $(TMPDIR) find -G OCR-D-SEG); \
|
||||
fgrep -q http://schema.primaresearch.org/PAGE/gts/pagecontent/2019-07-15 $(TMPDIR)/$$result && \
|
||||
fgrep -c -e TextRegion -e ImageRegion -e SeparatorRegion $(TMPDIR)/$$result
|
||||
ocrd-sbb-binarize -w $(TMPDIR) -I OCR-D-IMG -O OCR-D-BIN -P model $(CURDIR)
|
||||
ocrd-sbb-binarize -w $(TMPDIR) -I OCR-D-SEG -O OCR-D-SEG-BIN -P model $(CURDIR) -P operation_level region
|
||||
ocrd-sbb-binarize -w $(TMPDIR) -I OCR-D-IMG -O OCR-D-BIN -P model $(CURDIR)/default-2021-03-09
|
||||
ocrd-sbb-binarize -w $(TMPDIR) -I OCR-D-SEG -O OCR-D-SEG-BIN -P model $(CURDIR)/default-2021-03-09 -P operation_level region
|
||||
$(RM) -r $(TMPDIR)
|
||||
|
||||
# Run unit tests
|
||||
test: export EYNOLLAH_MODELS_DIR := $(CURDIR)
|
||||
test: export EYNOLLAH_MODELS=$(CURDIR)/models_eynollah
|
||||
test: export SBBBIN_MODELS=$(CURDIR)/default-2021-03-09
|
||||
test:
|
||||
$(PYTHON) -m pytest tests --durations=0 --continue-on-collection-errors $(PYTEST_ARGS)
|
||||
|
||||
|
|
@ -122,7 +117,7 @@ coverage:
|
|||
|
||||
# Build docker image
|
||||
docker:
|
||||
$(DOCKER) build \
|
||||
docker build \
|
||||
--build-arg DOCKER_BASE_IMAGE=$(DOCKER_BASE_IMAGE) \
|
||||
--build-arg VCS_REF=$$(git rev-parse --short HEAD) \
|
||||
--build-arg BUILD_DATE=$$(date -u +"%Y-%m-%dT%H:%M:%SZ") \
|
||||
|
|
|
|||
201
README.md
201
README.md
|
|
@ -1,33 +1,30 @@
|
|||
# Eynollah
|
||||
> Document Layout Analysis with Deep Learning and Heuristics
|
||||
|
||||
> Document Layout Analysis, Binarization and OCR with Deep Learning and Heuristics
|
||||
|
||||
[](https://pypi.python.org/pypi/eynollah)
|
||||
[](https://pypi.org/project/eynollah/)
|
||||
[](https://github.com/qurator-spk/eynollah/actions/workflows/test-eynollah.yml)
|
||||
[](https://github.com/qurator-spk/eynollah/actions/workflows/build-docker.yml)
|
||||
[](https://opensource.org/license/apache-2-0/)
|
||||
[](https://opensource.org/license/apache-2-0/)
|
||||
[](https://doi.org/10.1145/3604951.3605513)
|
||||
|
||||
|
||||
|
||||
## Features
|
||||
* Document layout analysis using pixelwise segmentation models with support for 10 segmentation classes:
|
||||
* Support for up to 10 segmentation classes:
|
||||
* background, [page border](https://ocr-d.de/en/gt-guidelines/trans/lyRand.html), [text region](https://ocr-d.de/en/gt-guidelines/trans/lytextregion.html#textregionen__textregion_), [text line](https://ocr-d.de/en/gt-guidelines/pagexml/pagecontent_xsd_Complex_Type_pc_TextLineType.html), [header](https://ocr-d.de/en/gt-guidelines/trans/lyUeberschrift.html), [image](https://ocr-d.de/en/gt-guidelines/trans/lyBildbereiche.html), [separator](https://ocr-d.de/en/gt-guidelines/trans/lySeparatoren.html), [marginalia](https://ocr-d.de/en/gt-guidelines/trans/lyMarginalie.html), [initial](https://ocr-d.de/en/gt-guidelines/trans/lyInitiale.html), [table](https://ocr-d.de/en/gt-guidelines/trans/lyTabellen.html)
|
||||
* Textline segmentation to bounding boxes or polygons (contours) including for curved lines and vertical text
|
||||
* Document image binarization with pixelwise segmentation or hybrid CNN-Transformer models
|
||||
* Text recognition (OCR) with CNN-RNN or TrOCR models
|
||||
* Detection of reading order (left-to-right or right-to-left) using heuristics or trainable models
|
||||
* Support for various image optimization operations:
|
||||
* cropping (border detection), binarization, deskewing, dewarping, scaling, enhancing, resizing
|
||||
* Text line segmentation to bounding boxes or polygons (contours) including for curved lines and vertical text
|
||||
* Detection of reading order (left-to-right or right-to-left)
|
||||
* Output in [PAGE-XML](https://github.com/PRImA-Research-Lab/PAGE-XML)
|
||||
* [OCR-D](https://github.com/qurator-spk/eynollah#use-as-ocr-d-processor) interface
|
||||
|
||||
:warning: Development is focused on achieving the best quality of results for a wide variety of historical
|
||||
documents using a combination of multiple deep learning models and heuristics; therefore processing can be slow.
|
||||
:warning: Development is currently focused on achieving the best possible quality of results for a wide variety of historical documents and therefore processing can be very slow. We aim to improve this, but contributions are welcome.
|
||||
|
||||
## Installation
|
||||
Python `3.8-3.11` with Tensorflow `<2.13` on Linux are currently supported.
|
||||
For (limited) GPU support the CUDA toolkit needs to be installed.
|
||||
A working config is CUDA `11.8` with cuDNN `8.6`.
|
||||
|
||||
For (limited) GPU support the CUDA toolkit needs to be installed.
|
||||
|
||||
You can either install from PyPI
|
||||
|
||||
|
|
@ -44,54 +41,19 @@ cd eynollah; pip install -e .
|
|||
|
||||
Alternatively, you can run `make install` or `make install-dev` for editable installation.
|
||||
|
||||
To also install the dependencies for the OCR engines:
|
||||
|
||||
```
|
||||
pip install "eynollah[OCR]"
|
||||
# or
|
||||
make install EXTRAS=OCR
|
||||
```
|
||||
|
||||
### Docker
|
||||
|
||||
Use
|
||||
|
||||
```
|
||||
docker pull ghcr.io/qurator-spk/eynollah:latest
|
||||
```
|
||||
|
||||
When using Eynollah with Docker, see [`docker.md`](https://github.com/qurator-spk/eynollah/tree/main/docs/docker.md).
|
||||
|
||||
## Models
|
||||
Pre-trained models can be downloaded from [qurator-data.de](https://qurator-data.de/eynollah/) or [huggingface](https://huggingface.co/SBB?search_models=eynollah).
|
||||
|
||||
Pretrained models can be downloaded from [Zenodo](https://zenodo.org/records/17194824) or [Hugging Face](https://huggingface.co/SBB?search_models=eynollah).
|
||||
For documentation on methods and models, have a look at [`models.md`](https://github.com/qurator-spk/eynollah/tree/main/docs/models.md).
|
||||
|
||||
For model documentation and model cards, see [`models.md`](https://github.com/qurator-spk/eynollah/tree/main/docs/models.md).
|
||||
|
||||
## Training
|
||||
|
||||
To train your own model with Eynollah, see [`train.md`](https://github.com/qurator-spk/eynollah/tree/main/docs/train.md) and use the tools in the [`train`](https://github.com/qurator-spk/eynollah/tree/main/train) folder.
|
||||
## Train
|
||||
In case you want to train your own model with Eynollah, have a look at [`train.md`](https://github.com/qurator-spk/eynollah/tree/main/docs/train.md).
|
||||
|
||||
## Usage
|
||||
|
||||
Eynollah supports five use cases:
|
||||
1. [layout analysis (segmentation)](#layout-analysis),
|
||||
2. [binarization](#binarization),
|
||||
3. [image enhancement](#image-enhancement),
|
||||
4. [text recognition (OCR)](#ocr), and
|
||||
5. [reading order detection](#reading-order-detection).
|
||||
|
||||
Some example outputs can be found in [`examples.md`](https://github.com/qurator-spk/eynollah/tree/main/docs/examples.md).
|
||||
|
||||
### Layout Analysis
|
||||
|
||||
The layout analysis module is responsible for detecting layout elements, identifying text lines, and determining reading
|
||||
order using heuristic methods or a [pretrained model](https://github.com/qurator-spk/eynollah#machine-based-reading-order).
|
||||
|
||||
The command-line interface for layout analysis can be called like this:
|
||||
The command-line interface can be called like this:
|
||||
|
||||
```sh
|
||||
eynollah layout \
|
||||
eynollah \
|
||||
-i <single image file> | -di <directory containing image files> \
|
||||
-o <output directory> \
|
||||
-m <directory containing model files> \
|
||||
|
|
@ -100,102 +62,57 @@ eynollah layout \
|
|||
|
||||
The following options can be used to further configure the processing:
|
||||
|
||||
| option | description |
|
||||
|-------------------|:--------------------------------------------------------------------------------------------|
|
||||
| `-fl` | full layout analysis including all steps and segmentation classes (recommended) |
|
||||
| `-tab` | apply table detection |
|
||||
| `-ae` | apply enhancement (the resulting image is saved to the output directory) |
|
||||
| `-as` | apply scaling |
|
||||
| `-cl` | apply contour detection for curved text lines instead of bounding boxes |
|
||||
| `-ib` | apply binarization (the resulting image is saved to the output directory) |
|
||||
| `-ep` | enable plotting (MUST always be used with `-sl`, `-sd`, `-sa`, `-si` or `-ae`) |
|
||||
| `-ho` | ignore headers for reading order dectection |
|
||||
| `-si <directory>` | save image regions detected to this directory |
|
||||
| `-sd <directory>` | save deskewed image to this directory |
|
||||
| `-sl <directory>` | save layout prediction as plot to this directory |
|
||||
| `-sp <directory>` | save cropped page image to this directory |
|
||||
| `-sa <directory>` | save all (plot, enhanced/binary image, layout) to this directory |
|
||||
| `-thart` | threshold of artifical class in the case of textline detection. The default value is 0.1 |
|
||||
| `-tharl` | threshold of artifical class in the case of layout detection. The default value is 0.1 |
|
||||
| `-ncu` | upper limit of columns in document image |
|
||||
| `-ncl` | lower limit of columns in document image |
|
||||
| `-slro` | skip layout detection and reading order |
|
||||
| `-romb` | apply machine based reading order detection |
|
||||
| `-ipe` | ignore page extraction |
|
||||
| option | description |
|
||||
|-------------------|:-------------------------------------------------------------------------------|
|
||||
| `-fl` | full layout analysis including all steps and segmentation classes |
|
||||
| `-light` | lighter and faster but simpler method for main region detection and deskewing |
|
||||
| `-tab` | apply table detection |
|
||||
| `-ae` | apply enhancement (the resulting image is saved to the output directory) |
|
||||
| `-as` | apply scaling |
|
||||
| `-cl` | apply contour detection for curved text lines instead of bounding boxes |
|
||||
| `-ib` | apply binarization (the resulting image is saved to the output directory) |
|
||||
| `-ep` | enable plotting (MUST always be used with `-sl`, `-sd`, `-sa`, `-si` or `-ae`) |
|
||||
| `-eoi` | extract only images to output directory (other processing will not be done) |
|
||||
| `-ho` | ignore headers for reading order dectection |
|
||||
| `-si <directory>` | save image regions detected to this directory |
|
||||
| `-sd <directory>` | save deskewed image to this directory |
|
||||
| `-sl <directory>` | save layout prediction as plot to this directory |
|
||||
| `-sp <directory>` | save cropped page image to this directory |
|
||||
| `-sa <directory>` | save all (plot, enhanced/binary image, layout) to this directory |
|
||||
|
||||
If no option is set, the tool performs layout detection of main regions (background, text, images, separators and marginals).
|
||||
The best output quality is produced when RGB images are used as input rather than greyscale or binarized images.
|
||||
|
||||
#### Use as OCR-D processor
|
||||
|
||||
Eynollah ships with a CLI interface to be used as [OCR-D](https://ocr-d.de) [processor](https://ocr-d.de/en/spec/cli),
|
||||
formally described in [`ocrd-tool.json`](https://github.com/qurator-spk/eynollah/tree/main/src/eynollah/ocrd-tool.json).
|
||||
|
||||
In this case, the source image file group with (preferably) RGB images should be used as input like this:
|
||||
|
||||
ocrd-eynollah-segment -I OCR-D-IMG -O OCR-D-SEG -P models 2022-04-05
|
||||
|
||||
|
||||
If no further option is set, the tool performs layout detection of main regions (background, text, images, separators
|
||||
and marginals).
|
||||
The best output quality is achieved when RGB images are used as input rather than greyscale or binarized images.
|
||||
|
||||
Additional documentation can be found in [`usage.md`](https://github.com/qurator-spk/eynollah/tree/main/docs/usage.md).
|
||||
|
||||
### Binarization
|
||||
|
||||
The binarization module performs document image binarization using pretrained pixelwise segmentation models.
|
||||
|
||||
The command-line interface for binarization can be called like this:
|
||||
|
||||
```sh
|
||||
eynollah binarization \
|
||||
-i <single image file> | -di <directory containing image files> \
|
||||
-o <output directory> \
|
||||
-m <directory containing model files>
|
||||
```
|
||||
|
||||
### Image Enhancement
|
||||
TODO
|
||||
|
||||
### OCR
|
||||
|
||||
The OCR module performs text recognition using either a CNN-RNN model or a Transformer model.
|
||||
|
||||
The command-line interface for OCR can be called like this:
|
||||
|
||||
```sh
|
||||
eynollah ocr \
|
||||
-i <single image file> | -di <directory containing image files> \
|
||||
-dx <directory of xmls> \
|
||||
-o <output directory> \
|
||||
-m <directory containing model files> | --model_name <path to specific model>
|
||||
```
|
||||
|
||||
The following options can be used to further configure the ocr processing:
|
||||
|
||||
| option | description |
|
||||
|-------------------|:-------------------------------------------------------------------------------------------|
|
||||
| `-dib` | directory of binarized images (file type must be '.png'), prediction with both RGB and bin |
|
||||
| `-doit` | directory for output images rendered with the predicted text |
|
||||
| `--model_name` | file path to use specific model for OCR |
|
||||
| `-trocr` | use transformer ocr model (otherwise cnn_rnn model is used) |
|
||||
| `-etit` | export textline images and text in xml to output dir (OCR training data) |
|
||||
| `-nmtc` | cropped textline images will not be masked with textline contour |
|
||||
| `-bs` | ocr inference batch size. Default batch size is 2 for trocr and 8 for cnn_rnn models |
|
||||
| `-ds_pref` | add an abbrevation of dataset name to generated training data |
|
||||
| `-min_conf` | minimum OCR confidence value. OCR with textline conf lower than this will be ignored |
|
||||
If the input file group is PAGE-XML (from a previous OCR-D workflow step), Eynollah behaves as follows:
|
||||
- existing regions are kept and ignored (i.e. in effect they might overlap segments from Eynollah results)
|
||||
- existing annotation (and respective `AlternativeImage`s) are partially _ignored_:
|
||||
- previous page frame detection (`cropped` images)
|
||||
- previous derotation (`deskewed` images)
|
||||
- previous thresholding (`binarized` images)
|
||||
- if the page-level image nevertheless deviates from the original (`@imageFilename`)
|
||||
(because some other preprocessing step was in effect like `denoised`), then
|
||||
the output PAGE-XML will be based on that as new top-level (`@imageFilename`)
|
||||
|
||||
|
||||
### Reading Order Detection
|
||||
Reading order detection can be performed either as part of layout analysis based on image input, or, currently under
|
||||
development, based on pre-existing layout analysis data in PAGE-XML format as input.
|
||||
ocrd-eynollah-segment -I OCR-D-XYZ -O OCR-D-SEG -P models 2022-04-05
|
||||
|
||||
The reading order detection module employs a pretrained model to identify the reading order from layouts represented in PAGE-XML files.
|
||||
Still, in general, it makes more sense to add other workflow steps **after** Eynollah.
|
||||
|
||||
The command-line interface for machine based reading order can be called like this:
|
||||
|
||||
```sh
|
||||
eynollah machine-based-reading-order \
|
||||
-i <single image file> | -di <directory containing image files> \
|
||||
-xml <xml file name> | -dx <directory containing xml files> \
|
||||
-m <path to directory containing model files> \
|
||||
-o <output directory>
|
||||
```
|
||||
|
||||
## Use as OCR-D processor
|
||||
|
||||
See [`ocrd.md`](https://github.com/qurator-spk/eynollah/tree/main/docs/ocrd.md).
|
||||
#### Additional documentation
|
||||
Please check the [wiki](https://github.com/qurator-spk/eynollah/wiki).
|
||||
|
||||
## How to cite
|
||||
If you find this tool useful in your work, please consider citing our paper:
|
||||
|
||||
```bibtex
|
||||
@inproceedings{hip23rezanezhad,
|
||||
|
|
|
|||
|
|
@ -1,43 +0,0 @@
|
|||
## Inference with Docker
|
||||
|
||||
docker pull ghcr.io/qurator-spk/eynollah:latest
|
||||
|
||||
### 1. ocrd resource manager
|
||||
(just once, to get the models and install them into a named volume for later re-use)
|
||||
|
||||
vol_models=ocrd-resources:/usr/local/share/ocrd-resources
|
||||
docker run --rm -v $vol_models ocrd/eynollah ocrd resmgr download ocrd-eynollah-segment default
|
||||
|
||||
Now, each time you want to use Eynollah, pass the same resources volume again.
|
||||
Also, bind-mount some data directory, e.g. current working directory $PWD (/data is default working directory in the container).
|
||||
|
||||
Either use standalone CLI (2) or OCR-D CLI (3):
|
||||
|
||||
### 2. standalone CLI
|
||||
(follow self-help, cf. readme)
|
||||
|
||||
docker run --rm -v $vol_models -v $PWD:/data ocrd/eynollah eynollah binarization --help
|
||||
docker run --rm -v $vol_models -v $PWD:/data ocrd/eynollah eynollah layout --help
|
||||
docker run --rm -v $vol_models -v $PWD:/data ocrd/eynollah eynollah ocr --help
|
||||
|
||||
### 3. OCR-D CLI
|
||||
(follow self-help, cf. readme and https://ocr-d.de/en/spec/cli)
|
||||
|
||||
docker run --rm -v $vol_models -v $PWD:/data ocrd/eynollah ocrd-eynollah-segment -h
|
||||
docker run --rm -v $vol_models -v $PWD:/data ocrd/eynollah ocrd-sbb-binarize -h
|
||||
|
||||
Alternatively, just "log in" to the container once and use the commands there:
|
||||
|
||||
docker run --rm -v $vol_models -v $PWD:/data -it ocrd/eynollah bash
|
||||
|
||||
## Training with Docker
|
||||
|
||||
Build the Docker training image
|
||||
|
||||
cd train
|
||||
docker build -t model-training .
|
||||
|
||||
Run the Docker training image
|
||||
|
||||
cd train
|
||||
docker run --gpus all -v $PWD:/entry_point_dir model-training
|
||||
|
|
@ -1,18 +0,0 @@
|
|||
# Examples
|
||||
|
||||
Example outputs of various Eynollah models
|
||||
|
||||
# Binarisation
|
||||
|
||||
<img src="https://user-images.githubusercontent.com/952378/63592437-e433e400-c5b1-11e9-9c2d-889c6e93d748.jpg" width="45%"><img src="https://user-images.githubusercontent.com/952378/63592435-e433e400-c5b1-11e9-88e4-3e441b61fa67.jpg" width="45%">
|
||||
<img src="https://user-images.githubusercontent.com/952378/63592440-e4cc7a80-c5b1-11e9-8964-2cd1b22c87be.jpg" width="45%"><img src="https://user-images.githubusercontent.com/952378/63592438-e4cc7a80-c5b1-11e9-86dc-a9e9f8555422.jpg" width="45%">
|
||||
|
||||
# Reading Order Detection
|
||||
|
||||
<img src="https://github.com/user-attachments/assets/42df2582-4579-415e-92f1-54858a02c830" alt="Input Image" width="45%">
|
||||
<img src="https://github.com/user-attachments/assets/77fc819e-6302-4fc9-967c-ee11d10d863e" alt="Output Image" width="45%">
|
||||
|
||||
# OCR
|
||||
|
||||
<img src="https://github.com/user-attachments/assets/71054636-51c6-4117-b3cf-361c5cda3528" alt="Input Image" width="45%"><img src="https://github.com/user-attachments/assets/cfb3ce38-007a-4037-b547-21324a7d56dd" alt="Output Image" width="45%">
|
||||
<img src="https://github.com/user-attachments/assets/343b2ed8-d818-4d4a-b301-f304cbbebfcd" alt="Input Image" width="45%"><img src="https://github.com/user-attachments/assets/accb5ba7-e37f-477e-84aa-92eafa0d136e" alt="Output Image" width="45%">
|
||||
|
|
@ -1,6 +1,5 @@
|
|||
# Models documentation
|
||||
|
||||
This suite of 15 models presents a document layout analysis (DLA) system for historical documents implemented by
|
||||
This suite of 14 models presents a document layout analysis (DLA) system for historical documents implemented by
|
||||
pixel-wise segmentation using a combination of a ResNet50 encoder with various U-Net decoders. In addition, heuristic
|
||||
methods are applied to detect marginals and to determine the reading order of text regions.
|
||||
|
||||
|
|
@ -18,14 +17,12 @@ Two Arabic/Persian terms form the name of the model suite: عين الله, whic
|
|||
|
||||
See the flowchart below for the different stages and how they interact:
|
||||
|
||||
<img width="810" height="691" alt="eynollah_flowchart" src="https://github.com/user-attachments/assets/42dd55bc-7b85-4b46-9afe-15ff712607f0" />
|
||||
|
||||
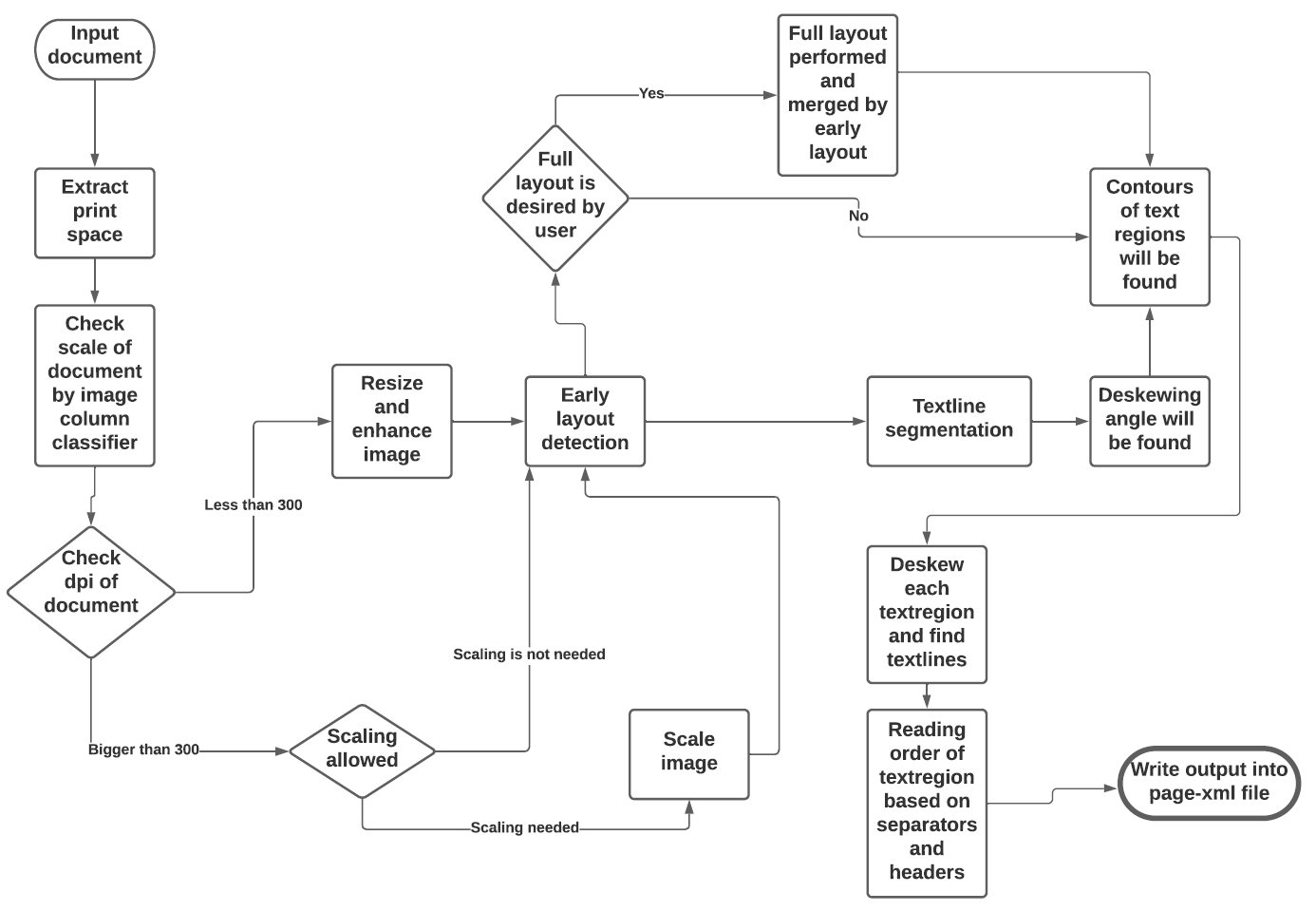
|
||||
|
||||
|
||||
## Models
|
||||
|
||||
### Image enhancement
|
||||
|
||||
Model card: [Image Enhancement](https://huggingface.co/SBB/eynollah-enhancement)
|
||||
|
||||
This model addresses image resolution, specifically targeting documents with suboptimal resolution. In instances where
|
||||
|
|
@ -33,14 +30,12 @@ the detection of document layout exhibits inadequate performance, the proposed e
|
|||
the quality and clarity of the images, thus facilitating enhanced visual interpretation and analysis.
|
||||
|
||||
### Page extraction / border detection
|
||||
|
||||
Model card: [Page Extraction/Border Detection](https://huggingface.co/SBB/eynollah-page-extraction)
|
||||
|
||||
A problem that can negatively affect OCR are black margins around a page caused by document scanning. A deep learning
|
||||
model helps to crop to the page borders by using a pixel-wise segmentation method.
|
||||
|
||||
### Column classification
|
||||
|
||||
Model card: [Column Classification](https://huggingface.co/SBB/eynollah-column-classifier)
|
||||
|
||||
This model is a trained classifier that recognizes the number of columns in a document by use of a training set with
|
||||
|
|
@ -48,7 +43,6 @@ manual classification of all documents into six classes with either one, two, th
|
|||
respectively.
|
||||
|
||||
### Binarization
|
||||
|
||||
Model card: [Binarization](https://huggingface.co/SBB/eynollah-binarization)
|
||||
|
||||
This model is designed to tackle the intricate task of document image binarization, which involves segmentation of the
|
||||
|
|
@ -58,7 +52,6 @@ capability of the model enables improved accuracy and reliability in subsequent
|
|||
enhanced document understanding and interpretation.
|
||||
|
||||
### Main region detection
|
||||
|
||||
Model card: [Main Region Detection](https://huggingface.co/SBB/eynollah-main-regions)
|
||||
|
||||
This model has employed a different set of labels, including an artificial class specifically designed to encompass the
|
||||
|
|
@ -68,7 +61,6 @@ during the inference phase. By incorporating this methodology, improved efficien
|
|||
model's ability to accurately identify and classify text regions within documents.
|
||||
|
||||
### Main region detection (with scaling augmentation)
|
||||
|
||||
Model card: [Main Region Detection (with scaling augmentation)](https://huggingface.co/SBB/eynollah-main-regions-aug-scaling)
|
||||
|
||||
Utilizing scaling augmentation, this model leverages the capability to effectively segment elements of extremely high or
|
||||
|
|
@ -77,14 +69,12 @@ categorizing and isolating such elements, thereby enhancing its overall performa
|
|||
documents with varying scale characteristics.
|
||||
|
||||
### Main region detection (with rotation augmentation)
|
||||
|
||||
Model card: [Main Region Detection (with rotation augmentation)](https://huggingface.co/SBB/eynollah-main-regions-aug-rotation)
|
||||
|
||||
This model takes advantage of rotation augmentation. This helps the tool to segment the vertical text regions in a
|
||||
robust way.
|
||||
|
||||
### Main region detection (ensembled)
|
||||
|
||||
Model card: [Main Region Detection (ensembled)](https://huggingface.co/SBB/eynollah-main-regions-ensembled)
|
||||
|
||||
The robustness of this model is attained through an ensembling technique that combines the weights from various epochs.
|
||||
|
|
@ -92,19 +82,16 @@ By employing this approach, the model achieves a high level of resilience and st
|
|||
strengths of multiple epochs to enhance its overall performance and deliver consistent and reliable results.
|
||||
|
||||
### Full region detection (1,2-column documents)
|
||||
|
||||
Model card: [Full Region Detection (1,2-column documents)](https://huggingface.co/SBB/eynollah-full-regions-1column)
|
||||
|
||||
This model deals with documents comprising of one and two columns.
|
||||
|
||||
### Full region detection (3,n-column documents)
|
||||
|
||||
Model card: [Full Region Detection (3,n-column documents)](https://huggingface.co/SBB/eynollah-full-regions-3pluscolumn)
|
||||
|
||||
This model is responsible for detecting headers and drop capitals in documents with three or more columns.
|
||||
|
||||
### Textline detection
|
||||
|
||||
Model card: [Textline Detection](https://huggingface.co/SBB/eynollah-textline)
|
||||
|
||||
The method for textline detection combines deep learning and heuristics. In the deep learning part, an image-to-image
|
||||
|
|
@ -119,7 +106,6 @@ segmentation is first deskewed and then the textlines are separated with the sam
|
|||
textline bounding boxes. Later, the strap is rotated back into its original orientation.
|
||||
|
||||
### Textline detection (light)
|
||||
|
||||
Model card: [Textline Detection Light (simpler but faster method)](https://huggingface.co/SBB/eynollah-textline_light)
|
||||
|
||||
The method for textline detection combines deep learning and heuristics. In the deep learning part, an image-to-image
|
||||
|
|
@ -133,7 +119,6 @@ enhancing the model's ability to accurately identify and delineate individual te
|
|||
eliminates the need for additional heuristics in extracting textline contours.
|
||||
|
||||
### Table detection
|
||||
|
||||
Model card: [Table Detection](https://huggingface.co/SBB/eynollah-tables)
|
||||
|
||||
The objective of this model is to perform table segmentation in historical document images. Due to the pixel-wise
|
||||
|
|
@ -143,84 +128,20 @@ effectively identify and delineate tables within the historical document images,
|
|||
enabling subsequent analysis and interpretation.
|
||||
|
||||
### Image detection
|
||||
|
||||
Model card: [Image Detection](https://huggingface.co/SBB/eynollah-image-extraction)
|
||||
|
||||
This model is used for the task of illustration detection only.
|
||||
|
||||
### Reading order detection
|
||||
|
||||
Model card: [Reading Order Detection]()
|
||||
|
||||
The model extracts the reading order of text regions from the layout by classifying pairwise relationships between them. A sorting algorithm then determines the overall reading sequence.
|
||||
|
||||
### OCR
|
||||
|
||||
We have trained three OCR models: two CNN-RNN–based models and one transformer-based TrOCR model. The CNN-RNN models are generally faster and provide better results in most cases, though their performance decreases with heavily degraded images. The TrOCR model, on the other hand, is computationally expensive and slower during inference, but it can possibly produce better results on strongly degraded images.
|
||||
|
||||
#### CNN-RNN model: model_eynollah_ocr_cnnrnn_20250805
|
||||
|
||||
This model is trained on data where most of the samples are in Fraktur german script.
|
||||
|
||||
| Dataset | Input | CER | WER |
|
||||
|-----------------------|:-------|:-----------|:----------|
|
||||
| OCR-D-GT-Archiveform | BIN | 0.02147 | 0.05685 |
|
||||
| OCR-D-GT-Archiveform | RGB | 0.01636 | 0.06285 |
|
||||
|
||||
#### CNN-RNN model: model_eynollah_ocr_cnnrnn_20250904 (Default)
|
||||
|
||||
Compared to the model_eynollah_ocr_cnnrnn_20250805 model, this model is trained on a larger proportion of Antiqua data and achieves superior performance.
|
||||
|
||||
| Dataset | Input | CER | WER |
|
||||
|-----------------------|:------------|:-----------|:----------|
|
||||
| OCR-D-GT-Archiveform | BIN | 0.01635 | 0.05410 |
|
||||
| OCR-D-GT-Archiveform | RGB | 0.01471 | 0.05813 |
|
||||
| BLN600 | RGB | 0.04409 | 0.08879 |
|
||||
| BLN600 | Enhanced | 0.03599 | 0.06244 |
|
||||
|
||||
|
||||
#### Transformer OCR model: model_eynollah_ocr_trocr_20250919
|
||||
|
||||
This transformer OCR model is trained on the same data as model_eynollah_ocr_trocr_20250919.
|
||||
|
||||
| Dataset | Input | CER | WER |
|
||||
|-----------------------|:------------|:-----------|:----------|
|
||||
| OCR-D-GT-Archiveform | BIN | 0.01841 | 0.05589 |
|
||||
| OCR-D-GT-Archiveform | RGB | 0.01552 | 0.06177 |
|
||||
| BLN600 | RGB | 0.06347 | 0.13853 |
|
||||
|
||||
##### Qualitative evaluation of the models
|
||||
|
||||
| <img width="1600" src="https://github.com/user-attachments/assets/120fec0c-c370-46a6-b132-b0af800607cf"> | <img width="1000" src="https://github.com/user-attachments/assets/d84e6819-0a2a-4b3a-bb7d-ceac941babc4"> | <img width="1000" src="https://github.com/user-attachments/assets/bdd27cdb-bbec-4223-9a86-de7a27c6d018"> | <img width="1000" src="https://github.com/user-attachments/assets/1a507c75-75de-4da3-9545-af3746b9a207"> |
|
||||
|:---:|:---:|:---:|:---:|
|
||||
| Image | cnnrnn_20250805 | cnnrnn_20250904 | trocr_20250919 |
|
||||
|
||||
|
||||
|
||||
| <img width="2000" src="https://github.com/user-attachments/assets/9bc13d48-2a92-45fc-88db-c07ffadba067"> | <img width="1000" src="https://github.com/user-attachments/assets/2b294aeb-1362-4d6e-b70f-8aeffd94c5e7"> | <img width="1000" src="https://github.com/user-attachments/assets/9911317e-632e-4e6a-8839-1fb7e783da11"> | <img width="1000" src="https://github.com/user-attachments/assets/2c5626d9-0d23-49d3-80f5-a95f629c9c76"> |
|
||||
|:---:|:---:|:---:|:---:|
|
||||
| Image | cnnrnn_20250805 | cnnrnn_20250904 | trocr_20250919 |
|
||||
|
||||
|
||||
| <img width="2000" src="https://github.com/user-attachments/assets/d54d8510-5c6a-4ab0-9ba7-f6ec4ad452c6"> | <img width="1000" src="https://github.com/user-attachments/assets/a418b25b-00dc-493a-b3a3-b325b9b0cb85"> | <img width="1000" src="https://github.com/user-attachments/assets/df6e2b9e-a821-4b4c-8868-0c765700c341"> | <img width="1000" src="https://github.com/user-attachments/assets/b90277f5-40f4-4c99-80a2-da400f7d3640"> |
|
||||
|:---:|:---:|:---:|:---:|
|
||||
| Image | cnnrnn_20250805 | cnnrnn_20250904 | trocr_20250919 |
|
||||
|
||||
|
||||
| <img width="2000" src="https://github.com/user-attachments/assets/7ec49211-099f-4c21-9e60-47bfdf21f1b6"> | <img width="1000" src="https://github.com/user-attachments/assets/00ef9785-8885-41b3-bf6e-21eab743df71"> | <img width="1000" src="https://github.com/user-attachments/assets/13eb9f62-4d5a-46dc-befc-b02eb4f31fc1"> | <img width="1000" src="https://github.com/user-attachments/assets/a5c078d1-6d15-4d12-9040-526d7063d459"> |
|
||||
|:---:|:---:|:---:|:---:|
|
||||
| Image | cnnrnn_20250805 | cnnrnn_20250904 | trocr_20250919 |
|
||||
|
||||
|
||||
TODO
|
||||
|
||||
## Heuristic methods
|
||||
|
||||
Additionally, some heuristic methods are employed to further improve the model predictions:
|
||||
|
||||
* After border detection, the largest contour is determined by a bounding box, and the image cropped to these coordinates.
|
||||
* Unlike the non-light version, where the image is scaled up to help the model better detect the background spaces between text regions, the light version uses down-scaled images. In this case, introducing an artificial class along the boundaries of text regions and text lines has helped to isolate and separate the text regions more effectively.
|
||||
* For text region detection, the image is scaled up to make it easier for the model to detect background space between text regions.
|
||||
* A minimum area is defined for text regions in relation to the overall image dimensions, so that very small regions that are noise can be filtered out.
|
||||
* In the non-light version, deskewing is applied at the text-region level (since regions may have different degrees of skew) to improve text-line segmentation results. In contrast, the light version performs deskewing only at the page level to enhance margin detection and heuristic reading-order estimation.
|
||||
* After deskewing, a calculation of the pixel distribution on the X-axis allows the separation of textlines (foreground) and background pixels (only in non-light version).
|
||||
* Finally, using the derived coordinates, bounding boxes are determined for each textline (only in non-light version).
|
||||
* As mentioned above, the reading order can be determined using a model; however, this approach is computationally expensive, time-consuming, and less accurate due to the limited amount of ground-truth data available for training. Therefore, our tool uses a heuristic reading-order detection method as the default. The heuristic approach relies on headers and separators to determine the reading order of text regions.
|
||||
* Deskewing is applied on the text region level (due to regions having different degrees of skew) in order to improve the textline segmentation result.
|
||||
* After deskewing, a calculation of the pixel distribution on the X-axis allows the separation of textlines (foreground) and background pixels.
|
||||
* Finally, using the derived coordinates, bounding boxes are determined for each textline.
|
||||
|
|
|
|||
26
docs/ocrd.md
26
docs/ocrd.md
|
|
@ -1,26 +0,0 @@
|
|||
## Use as OCR-D processor
|
||||
|
||||
Eynollah ships with a CLI interface to be used as [OCR-D](https://ocr-d.de) [processor](https://ocr-d.de/en/spec/cli),
|
||||
formally described in [`ocrd-tool.json`](https://github.com/qurator-spk/eynollah/tree/main/src/eynollah/ocrd-tool.json).
|
||||
|
||||
When using Eynollah in OCR-D, the source image file group with (preferably) RGB images should be used as input like this:
|
||||
|
||||
ocrd-eynollah-segment -I OCR-D-IMG -O OCR-D-SEG -P models eynollah_layout_v0_5_0
|
||||
|
||||
If the input file group is PAGE-XML (from a previous OCR-D workflow step), Eynollah behaves as follows:
|
||||
- existing regions are kept and ignored (i.e. in effect they might overlap segments from Eynollah results)
|
||||
- existing annotation (and respective `AlternativeImage`s) are partially _ignored_:
|
||||
- previous page frame detection (`cropped` images)
|
||||
- previous derotation (`deskewed` images)
|
||||
- previous thresholding (`binarized` images)
|
||||
- if the page-level image nevertheless deviates from the original (`@imageFilename`)
|
||||
(because some other preprocessing step was in effect like `denoised`), then
|
||||
the output PAGE-XML will be based on that as new top-level (`@imageFilename`)
|
||||
|
||||
ocrd-eynollah-segment -I OCR-D-XYZ -O OCR-D-SEG -P models eynollah_layout_v0_5_0
|
||||
|
||||
In general, it makes more sense to add other workflow steps **after** Eynollah.
|
||||
|
||||
There is also an OCR-D processor for binarization:
|
||||
|
||||
ocrd-sbb-binarize -I OCR-D-IMG -O OCR-D-BIN -P models default-2021-03-09
|
||||
407
docs/train.md
407
docs/train.md
|
|
@ -1,93 +1,38 @@
|
|||
# Prerequisistes
|
||||
|
||||
## 1. Install Eynollah with training dependencies
|
||||
|
||||
Clone the repository and install eynollah along with the dependencies necessary for training:
|
||||
|
||||
```sh
|
||||
git clone https://github.com/qurator-spk/eynollah
|
||||
cd eynollah
|
||||
pip install '.[training]'
|
||||
```
|
||||
|
||||
## 2. Pretrained encoder
|
||||
|
||||
Download our pretrained weights and add them to a `train/pretrained_model` folder:
|
||||
|
||||
```sh
|
||||
cd train
|
||||
wget -O pretrained_model.tar.gz https://zenodo.org/records/17243320/files/pretrained_model_v0_5_1.tar.gz?download=1
|
||||
tar xf pretrained_model.tar.gz
|
||||
```
|
||||
|
||||
## 3. Example data
|
||||
|
||||
### Binarization
|
||||
A small sample of training data for binarization experiment can be found on [Zenodo](https://zenodo.org/records/17243320/files/training_data_sample_binarization_v0_5_1.tar.gz?download=1),
|
||||
which contains `images` and `labels` folders.
|
||||
|
||||
## 4. Helpful tools
|
||||
|
||||
* [`pagexml2img`](https://github.com/qurator-spk/page2img)
|
||||
> Tool to extract 2-D or 3-D RGB images from PAGE-XML data. In the former case, the output will be 1 2-D image array which each class has filled with a pixel value. In the case of a 3-D RGB image,
|
||||
each class will be defined with a RGB value and beside images, a text file of classes will also be produced.
|
||||
* [`cocoSegmentationToPng`](https://github.com/nightrome/cocostuffapi/blob/17acf33aef3c6cc2d6aca46dcf084266c2778cf0/PythonAPI/pycocotools/cocostuffhelper.py#L130)
|
||||
> Convert COCO GT or results for a single image to a segmentation map and write it to disk.
|
||||
* [`ocrd-segment-extract-pages`](https://github.com/OCR-D/ocrd_segment/blob/master/ocrd_segment/extract_pages.py)
|
||||
> Extract region classes and their colours in mask (pseg) images. Allows the color map as free dict parameter, and comes with a default that mimics PageViewer's coloring for quick debugging; it also warns when regions do overlap.
|
||||
|
||||
# Training documentation
|
||||
This aims to assist users in preparing training datasets, training models, and performing inference with trained models.
|
||||
We cover various use cases including pixel-wise segmentation, image classification, image enhancement, and machine-based
|
||||
reading order detection. For each use case, we provide guidance on how to generate the corresponding training dataset.
|
||||
|
||||
This document aims to assist users in preparing training datasets, training models, and
|
||||
performing inference with trained models. We cover various use cases including
|
||||
pixel-wise segmentation, image classification, image enhancement, and
|
||||
machine-based reading order detection. For each use case, we provide guidance
|
||||
on how to generate the corresponding training dataset.
|
||||
|
||||
The following three tasks can all be accomplished using the code in the
|
||||
[`train`](https://github.com/qurator-spk/eynollah/tree/main/train) directory:
|
||||
The following three tasks can all be accomplished using the code in the
|
||||
[`train`](https://github.com/qurator-spk/sbb_pixelwise_segmentation/tree/unifying-training-models) directory:
|
||||
|
||||
* generate training dataset
|
||||
* train a model
|
||||
* inference with the trained model
|
||||
|
||||
## Training, evaluation and output
|
||||
|
||||
The train and evaluation folders should contain subfolders of `images` and `labels`.
|
||||
|
||||
The output folder should be an empty folder where the output model will be written to.
|
||||
|
||||
## Generate training dataset
|
||||
The script `generate_gt_for_training.py` is used for generating training datasets. As the results of the following
|
||||
command demonstrates, the dataset generator provides three different commands:
|
||||
|
||||
The script `generate_gt_for_training.py` is used for generating training datasets. As the results of the following
|
||||
command demonstrates, the dataset generator provides several subcommands:
|
||||
`python generate_gt_for_training.py --help`
|
||||
|
||||
```sh
|
||||
eynollah-training generate-gt --help
|
||||
```
|
||||
|
||||
The three most important subcommands are:
|
||||
These three commands are:
|
||||
|
||||
* image-enhancement
|
||||
* machine-based-reading-order
|
||||
* pagexml2label
|
||||
|
||||
### image-enhancement
|
||||
|
||||
Generating a training dataset for image enhancement is quite straightforward. All that is needed is a set of
|
||||
Generating a training dataset for image enhancement is quite straightforward. All that is needed is a set of
|
||||
high-resolution images. The training dataset can then be generated using the following command:
|
||||
|
||||
```sh
|
||||
eynollah-training image-enhancement \
|
||||
-dis "dir of high resolution images" \
|
||||
-dois "dir where degraded images will be written" \
|
||||
-dols "dir where the corresponding high resolution image will be written as label" \
|
||||
-scs "degrading scales json file"
|
||||
```
|
||||
`python generate_gt_for_training.py image-enhancement -dis "dir of high resolution images" -dois "dir where degraded
|
||||
images will be written" -dols "dir where the corresponding high resolution image will be written as label" -scs
|
||||
"degrading scales json file"`
|
||||
|
||||
The scales JSON file is a dictionary with a key named `scales` and values representing scales smaller than 1. Images are
|
||||
downscaled based on these scales and then upscaled again to their original size. This process causes the images to lose
|
||||
resolution at different scales. The degraded images are used as input images, and the original high-resolution images
|
||||
The scales JSON file is a dictionary with a key named 'scales' and values representing scales smaller than 1. Images are
|
||||
downscaled based on these scales and then upscaled again to their original size. This process causes the images to lose
|
||||
resolution at different scales. The degraded images are used as input images, and the original high-resolution images
|
||||
serve as labels. The enhancement model can be trained with this generated dataset. The scales JSON file looks like this:
|
||||
|
||||
```yaml
|
||||
|
|
@ -97,40 +42,31 @@ serve as labels. The enhancement model can be trained with this generated datase
|
|||
```
|
||||
|
||||
### machine-based-reading-order
|
||||
|
||||
For machine-based reading order, we aim to determine the reading priority between two sets of text regions. The model's
|
||||
input is a three-channel image: the first and last channels contain information about each of the two text regions,
|
||||
while the middle channel encodes prominent layout elements necessary for reading order, such as separators and headers.
|
||||
To generate the training dataset, our script requires a page XML file that specifies the image layout with the correct
|
||||
For machine-based reading order, we aim to determine the reading priority between two sets of text regions. The model's
|
||||
input is a three-channel image: the first and last channels contain information about each of the two text regions,
|
||||
while the middle channel encodes prominent layout elements necessary for reading order, such as separators and headers.
|
||||
To generate the training dataset, our script requires a page XML file that specifies the image layout with the correct
|
||||
reading order.
|
||||
|
||||
For output images, it is necessary to specify the width and height. Additionally, a minimum text region size can be set
|
||||
to filter out regions smaller than this minimum size. This minimum size is defined as the ratio of the text region area
|
||||
For output images, it is necessary to specify the width and height. Additionally, a minimum text region size can be set
|
||||
to filter out regions smaller than this minimum size. This minimum size is defined as the ratio of the text region area
|
||||
to the image area, with a default value of zero. To run the dataset generator, use the following command:
|
||||
|
||||
```shell
|
||||
eynollah-training generate-gt machine-based-reading-order \
|
||||
-dx "dir of GT xml files" \
|
||||
-domi "dir where output images will be written" \
|
||||
"" -docl "dir where the labels will be written" \
|
||||
-ih "height" \
|
||||
-iw "width" \
|
||||
-min "min area ratio"
|
||||
```
|
||||
`python generate_gt_for_training.py machine-based-reading-order -dx "dir of GT xml files" -domi "dir where output images
|
||||
will be written" -docl "dir where the labels will be written" -ih "height" -iw "width" -min "min area ratio"`
|
||||
|
||||
### pagexml2label
|
||||
|
||||
pagexml2label is designed to generate labels from GT page XML files for various pixel-wise segmentation use cases,
|
||||
including 'layout,' 'textline,' 'printspace,' 'glyph,' and 'word' segmentation.
|
||||
To train a pixel-wise segmentation model, we require images along with their corresponding labels. Our training script
|
||||
expects a PNG image where each pixel corresponds to a label, represented by an integer. The background is always labeled
|
||||
as zero, while other elements are assigned different integers. For instance, if we have ground truth data with four
|
||||
To train a pixel-wise segmentation model, we require images along with their corresponding labels. Our training script
|
||||
expects a PNG image where each pixel corresponds to a label, represented by an integer. The background is always labeled
|
||||
as zero, while other elements are assigned different integers. For instance, if we have ground truth data with four
|
||||
elements including the background, the classes would be labeled as 0, 1, 2, and 3 respectively.
|
||||
|
||||
In binary segmentation scenarios such as textline or page extraction, the background is encoded as 0, and the desired
|
||||
In binary segmentation scenarios such as textline or page extraction, the background is encoded as 0, and the desired
|
||||
element is automatically encoded as 1 in the PNG label.
|
||||
|
||||
To specify the desired use case and the elements to be extracted in the PNG labels, a custom JSON file can be passed.
|
||||
To specify the desired use case and the elements to be extracted in the PNG labels, a custom JSON file can be passed.
|
||||
For example, in the case of 'textline' detection, the JSON file would resemble this:
|
||||
|
||||
```yaml
|
||||
|
|
@ -164,35 +100,31 @@ A possible custom config json file for layout segmentation where the "printspace
|
|||
}
|
||||
```
|
||||
|
||||
For the layout use case, it is beneficial to first understand the structure of the page XML file and its elements.
|
||||
In a given image, the annotations of elements are recorded in a page XML file, including their contours and classes.
|
||||
For an image document, the known regions are 'textregion', 'separatorregion', 'imageregion', 'graphicregion',
|
||||
For the layout use case, it is beneficial to first understand the structure of the page XML file and its elements.
|
||||
In a given image, the annotations of elements are recorded in a page XML file, including their contours and classes.
|
||||
For an image document, the known regions are 'textregion', 'separatorregion', 'imageregion', 'graphicregion',
|
||||
'noiseregion', and 'tableregion'.
|
||||
|
||||
Text regions and graphic regions also have their own specific types. The known types for text regions are 'paragraph',
|
||||
'header', 'heading', 'marginalia', 'drop-capital', 'footnote', 'footnote-continued', 'signature-mark', 'page-number',
|
||||
and 'catch-word'. The known types for graphic regions are 'handwritten-annotation', 'decoration', 'stamp', and
|
||||
Text regions and graphic regions also have their own specific types. The known types for text regions are 'paragraph',
|
||||
'header', 'heading', 'marginalia', 'drop-capital', 'footnote', 'footnote-continued', 'signature-mark', 'page-number',
|
||||
and 'catch-word'. The known types for graphic regions are 'handwritten-annotation', 'decoration', 'stamp', and
|
||||
'signature'.
|
||||
Since we don't know all types of text and graphic regions, unknown cases can arise. To handle these, we have defined
|
||||
two additional types, "rest_as_paragraph" and "rest_as_decoration", to ensure that no unknown types are missed.
|
||||
Since we don't know all types of text and graphic regions, unknown cases can arise. To handle these, we have defined
|
||||
two additional types, "rest_as_paragraph" and "rest_as_decoration", to ensure that no unknown types are missed.
|
||||
This way, users can extract all known types from the labels and be confident that no unknown types are overlooked.
|
||||
|
||||
In the custom JSON file shown above, "header" and "heading" are extracted as the same class, while "marginalia" is shown
|
||||
as a different class. All other text region types, including "drop-capital," are grouped into the same class. For the
|
||||
graphic region, "stamp" has its own class, while all other types are classified together. "Image region" and "separator
|
||||
region" are also present in the label. However, other regions like "noise region" and "table region" will not be
|
||||
In the custom JSON file shown above, "header" and "heading" are extracted as the same class, while "marginalia" is shown
|
||||
as a different class. All other text region types, including "drop-capital," are grouped into the same class. For the
|
||||
graphic region, "stamp" has its own class, while all other types are classified together. "Image region" and "separator
|
||||
region" are also present in the label. However, other regions like "noise region" and "table region" will not be
|
||||
included in the label PNG file, even if they have information in the page XML files, as we chose not to include them.
|
||||
|
||||
```sh
|
||||
eynollah-training generate-gt pagexml2label \
|
||||
-dx "dir of GT xml files" \
|
||||
-do "dir where output label png files will be written" \
|
||||
-cfg "custom config json file" \
|
||||
-to "output type which has 2d and 3d. 2d is used for training and 3d is just to visualise the labels"
|
||||
```
|
||||
`python generate_gt_for_training.py pagexml2label -dx "dir of GT xml files" -do "dir where output label png files will
|
||||
be written" -cfg "custom config json file" -to "output type which has 2d and 3d. 2d is used for training and 3d is just
|
||||
to visualise the labels" "`
|
||||
|
||||
We have also defined an artificial class that can be added to the boundary of text region types or text lines. This key
|
||||
is called "artificial_class_on_boundary." If users want to apply this to certain text regions in the layout use case,
|
||||
We have also defined an artificial class that can be added to the boundary of text region types or text lines. This key
|
||||
is called "artificial_class_on_boundary." If users want to apply this to certain text regions in the layout use case,
|
||||
the example JSON config file should look like this:
|
||||
|
||||
```yaml
|
||||
|
|
@ -215,13 +147,13 @@ the example JSON config file should look like this:
|
|||
}
|
||||
```
|
||||
|
||||
This implies that the artificial class label, denoted by 7, will be present on PNG files and will only be added to the
|
||||
This implies that the artificial class label, denoted by 7, will be present on PNG files and will only be added to the
|
||||
elements labeled as "paragraph," "header," "heading," and "marginalia."
|
||||
|
||||
For "textline", "word", and "glyph", the artificial class on the boundaries will be activated only if the
|
||||
"artificial_class_label" key is specified in the config file. Its value should be set as 2 since these elements
|
||||
represent binary cases. For example, if the background and textline are denoted as 0 and 1 respectively, then the
|
||||
artificial class should be assigned the value 2. The example JSON config file should look like this for "textline" use
|
||||
For "textline", "word", and "glyph", the artificial class on the boundaries will be activated only if the
|
||||
"artificial_class_label" key is specified in the config file. Its value should be set as 2 since these elements
|
||||
represent binary cases. For example, if the background and textline are denoted as 0 and 1 respectively, then the
|
||||
artificial class should be assigned the value 2. The example JSON config file should look like this for "textline" use
|
||||
case:
|
||||
|
||||
```yaml
|
||||
|
|
@ -231,32 +163,25 @@ case:
|
|||
}
|
||||
```
|
||||
|
||||
If the coordinates of "PrintSpace" or "Border" are present in the page XML ground truth files, and the user wishes to
|
||||
crop only the print space area, this can be achieved by activating the "-ps" argument. However, it should be noted that
|
||||
in this scenario, since cropping will be applied to the label files, the directory of the original images must be
|
||||
provided to ensure that they are cropped in sync with the labels. This ensures that the correct images and labels
|
||||
If the coordinates of "PrintSpace" or "Border" are present in the page XML ground truth files, and the user wishes to
|
||||
crop only the print space area, this can be achieved by activating the "-ps" argument. However, it should be noted that
|
||||
in this scenario, since cropping will be applied to the label files, the directory of the original images must be
|
||||
provided to ensure that they are cropped in sync with the labels. This ensures that the correct images and labels
|
||||
required for training are obtained. The command should resemble the following:
|
||||
|
||||
```sh
|
||||
eynollah-training generate-gt pagexml2label \
|
||||
-dx "dir of GT xml files" \
|
||||
-do "dir where output label png files will be written" \
|
||||
-cfg "custom config json file" \
|
||||
-to "output type which has 2d and 3d. 2d is used for training and 3d is just to visualise the labels" \
|
||||
-ps \
|
||||
-di "dir where the org images are located" \
|
||||
-doi "dir where the cropped output images will be written"
|
||||
```
|
||||
`python generate_gt_for_training.py pagexml2label -dx "dir of GT xml files" -do "dir where output label png files will
|
||||
be written" -cfg "custom config json file" -to "output type which has 2d and 3d. 2d is used for training and 3d is just
|
||||
to visualise the labels" -ps -di "dir where the org images are located" -doi "dir where the cropped output images will
|
||||
be written" `
|
||||
|
||||
## Train a model
|
||||
|
||||
### classification
|
||||
|
||||
For the classification use case, we haven't provided a ground truth generator, as it's unnecessary. For classification,
|
||||
all we require is a training directory with subdirectories, each containing images of its respective classes. We need
|
||||
separate directories for training and evaluation, and the class names (subdirectories) must be consistent across both
|
||||
directories. Additionally, the class names should be specified in the config JSON file, as shown in the following
|
||||
example. If, for instance, we aim to classify "apple" and "orange," with a total of 2 classes, the
|
||||
For the classification use case, we haven't provided a ground truth generator, as it's unnecessary. For classification,
|
||||
all we require is a training directory with subdirectories, each containing images of its respective classes. We need
|
||||
separate directories for training and evaluation, and the class names (subdirectories) must be consistent across both
|
||||
directories. Additionally, the class names should be specified in the config JSON file, as shown in the following
|
||||
example. If, for instance, we aim to classify "apple" and "orange," with a total of 2 classes, the
|
||||
"classification_classes_name" key in the config file should appear as follows:
|
||||
|
||||
```yaml
|
||||
|
|
@ -281,7 +206,7 @@ example. If, for instance, we aim to classify "apple" and "orange," with a total
|
|||
|
||||
The "dir_train" should be like this:
|
||||
|
||||
```
|
||||
```
|
||||
.
|
||||
└── train # train directory
|
||||
├── apple # directory of images for apple class
|
||||
|
|
@ -290,7 +215,7 @@ The "dir_train" should be like this:
|
|||
|
||||
And the "dir_eval" the same structure as train directory:
|
||||
|
||||
```
|
||||
```
|
||||
.
|
||||
└── eval # evaluation directory
|
||||
├── apple # directory of images for apple class
|
||||
|
|
@ -300,13 +225,11 @@ And the "dir_eval" the same structure as train directory:
|
|||
|
||||
The classification model can be trained using the following command line:
|
||||
|
||||
```sh
|
||||
eynollah-training train with config_classification.json
|
||||
```
|
||||
`python train.py with config_classification.json`
|
||||
|
||||
As evident in the example JSON file above, for classification, we utilize a "f1_threshold_classification" parameter.
|
||||
This parameter is employed to gather all models with an evaluation f1 score surpassing this threshold. Subsequently,
|
||||
an ensemble of these model weights is executed, and a model is saved in the output directory as "model_ens_avg".
|
||||
As evident in the example JSON file above, for classification, we utilize a "f1_threshold_classification" parameter.
|
||||
This parameter is employed to gather all models with an evaluation f1 score surpassing this threshold. Subsequently,
|
||||
an ensemble of these model weights is executed, and a model is saved in the output directory as "model_ens_avg".
|
||||
Additionally, the weight of the best model based on the evaluation f1 score is saved as "model_best".
|
||||
|
||||
### reading order
|
||||
|
|
@ -353,64 +276,58 @@ The classification model can be trained like the classification case command lin
|
|||
### Segmentation (Textline, Binarization, Page extraction and layout) and enhancement
|
||||
|
||||
#### Parameter configuration for segmentation or enhancement usecases
|
||||
|
||||
The following parameter configuration can be applied to all segmentation use cases and enhancements. The augmentation,
|
||||
its sub-parameters, and continued training are defined only for segmentation use cases and enhancements, not for
|
||||
The following parameter configuration can be applied to all segmentation use cases and enhancements. The augmentation,
|
||||
its sub-parameters, and continued training are defined only for segmentation use cases and enhancements, not for
|
||||
classification and machine-based reading order, as you can see in their example config files.
|
||||
|
||||
* `backbone_type`: For segmentation tasks (such as text line, binarization, and layout detection) and enhancement, we
|
||||
offer two backbone options: a "nontransformer" and a "transformer" backbone. For the "transformer" backbone, we first
|
||||
apply a CNN followed by a transformer. In contrast, the "nontransformer" backbone utilizes only a CNN ResNet-50.
|
||||
* `task`: The task parameter can have values such as "segmentation", "enhancement", "classification", and "reading_order".
|
||||
* `patches`: If you want to break input images into smaller patches (input size of the model) you need to set this
|
||||
* parameter to `true`. In the case that the model should see the image once, like page extraction, patches should be
|
||||
set to ``false``.
|
||||
* `n_batch`: Number of batches at each iteration.
|
||||
* `n_classes`: Number of classes. In the case of binary classification this should be 2. In the case of reading_order it
|
||||
should set to 1. And for the case of layout detection just the unique number of classes should be given.
|
||||
* `n_epochs`: Number of epochs.
|
||||
* `input_height`: This indicates the height of model's input.
|
||||
* `input_width`: This indicates the width of model's input.
|
||||
* `weight_decay`: Weight decay of l2 regularization of model layers.
|
||||
* `pretraining`: Set to `true` to load pretrained weights of ResNet50 encoder. The downloaded weights should be saved
|
||||
in a folder named "pretrained_model" in the same directory of "train.py" script.
|
||||
* `augmentation`: If you want to apply any kind of augmentation this parameter should first set to `true`.
|
||||
* `flip_aug`: If `true`, different types of filp will be applied on image. Type of flips is given with "flip_index" parameter.
|
||||
* `blur_aug`: If `true`, different types of blurring will be applied on image. Type of blurrings is given with "blur_k" parameter.
|
||||
* `scaling`: If `true`, scaling will be applied on image. Scale of scaling is given with "scales" parameter.
|
||||
* `degrading`: If `true`, degrading will be applied to the image. The amount of degrading is defined with "degrade_scales" parameter.
|
||||
* `brightening`: If `true`, brightening will be applied to the image. The amount of brightening is defined with "brightness" parameter.
|
||||
* `rotation_not_90`: If `true`, rotation (not 90 degree) will be applied on image. Rotation angles are given with "thetha" parameter.
|
||||
* `rotation`: If `true`, 90 degree rotation will be applied on image.
|
||||
* `binarization`: If `true`,Otsu thresholding will be applied to augment the input data with binarized images.
|
||||
* `scaling_bluring`: If `true`, combination of scaling and blurring will be applied on image.
|
||||
* `scaling_binarization`: If `true`, combination of scaling and binarization will be applied on image.
|
||||
* `scaling_flip`: If `true`, combination of scaling and flip will be applied on image.
|
||||
* `flip_index`: Type of flips.
|
||||
* `blur_k`: Type of blurrings.
|
||||
* `scales`: Scales of scaling.
|
||||
* `brightness`: The amount of brightenings.
|
||||
* `thetha`: Rotation angles.
|
||||
* `degrade_scales`: The amount of degradings.
|
||||
* `continue_training`: If `true`, it means that you have already trained a model and you would like to continue the
|
||||
training. So it is needed to providethe dir of trained model with "dir_of_start_model" and index for naming
|
||||
themodels. For example if you have already trained for 3 epochs then your lastindex is 2 and if you want to continue
|
||||
from model_1.h5, you can set `index_start` to 3 to start naming model with index 3.
|
||||
* `weighted_loss`: If `true`, this means that you want to apply weighted categorical_crossentropy as loss fucntion. Be carefull if you set to `true`the parameter "is_loss_soft_dice" should be ``false``
|
||||
* `data_is_provided`: If you have already provided the input data you can set this to `true`. Be sure that the train
|
||||
and eval data are in"dir_output".Since when once we provide training data we resize and augmentthem and then wewrite
|
||||
them in sub-directories train and eval in "dir_output".
|
||||
* `dir_train`: This is the directory of "images" and "labels" (dir_train should include two subdirectories with names of images and labels ) for raw images and labels. Namely they are not prepared (not resized and not augmented) yet for training the model. When we run this tool these raw data will be transformed to suitable size needed for the model and they will be written in "dir_output" in train and eval directories. Each of train and eval include "images" and "labels" sub-directories.
|
||||
* `index_start`: Starting index for saved models in the case that "continue_training" is `true`.
|
||||
* `dir_of_start_model`: Directory containing pretrained model to continue training the model in the case that "continue_training" is `true`.
|
||||
* `transformer_num_patches_xy`: Number of patches for vision transformer in x and y direction respectively.
|
||||
* `transformer_patchsize_x`: Patch size of vision transformer patches in x direction.
|
||||
* `transformer_patchsize_y`: Patch size of vision transformer patches in y direction.
|
||||
* `transformer_projection_dim`: Transformer projection dimension. Default value is 64.
|
||||
* `transformer_mlp_head_units`: Transformer Multilayer Perceptron (MLP) head units. Default value is [128, 64].
|
||||
* `transformer_layers`: transformer layers. Default value is 8.
|
||||
* `transformer_num_heads`: Transformer number of heads. Default value is 4.
|
||||
* `transformer_cnn_first`: We have two types of vision transformers. In one type, a CNN is applied first, followed by a transformer. In the other type, this order is reversed. If transformer_cnn_first is true, it means the CNN will be applied before the transformer. Default value is true.
|
||||
* backbone_type: For segmentation tasks (such as text line, binarization, and layout detection) and enhancement, we
|
||||
* offer two backbone options: a "nontransformer" and a "transformer" backbone. For the "transformer" backbone, we first
|
||||
* apply a CNN followed by a transformer. In contrast, the "nontransformer" backbone utilizes only a CNN ResNet-50.
|
||||
* task : The task parameter can have values such as "segmentation", "enhancement", "classification", and "reading_order".
|
||||
* patches: If you want to break input images into smaller patches (input size of the model) you need to set this
|
||||
* parameter to ``true``. In the case that the model should see the image once, like page extraction, patches should be
|
||||
* set to ``false``.
|
||||
* n_batch: Number of batches at each iteration.
|
||||
* n_classes: Number of classes. In the case of binary classification this should be 2. In the case of reading_order it
|
||||
* should set to 1. And for the case of layout detection just the unique number of classes should be given.
|
||||
* n_epochs: Number of epochs.
|
||||
* input_height: This indicates the height of model's input.
|
||||
* input_width: This indicates the width of model's input.
|
||||
* weight_decay: Weight decay of l2 regularization of model layers.
|
||||
* pretraining: Set to ``true`` to load pretrained weights of ResNet50 encoder. The downloaded weights should be saved
|
||||
* in a folder named "pretrained_model" in the same directory of "train.py" script.
|
||||
* augmentation: If you want to apply any kind of augmentation this parameter should first set to ``true``.
|
||||
* flip_aug: If ``true``, different types of filp will be applied on image. Type of flips is given with "flip_index" parameter.
|
||||
* blur_aug: If ``true``, different types of blurring will be applied on image. Type of blurrings is given with "blur_k" parameter.
|
||||
* scaling: If ``true``, scaling will be applied on image. Scale of scaling is given with "scales" parameter.
|
||||
* degrading: If ``true``, degrading will be applied to the image. The amount of degrading is defined with "degrade_scales" parameter.
|
||||
* brightening: If ``true``, brightening will be applied to the image. The amount of brightening is defined with "brightness" parameter.
|
||||
* rotation_not_90: If ``true``, rotation (not 90 degree) will be applied on image. Rotation angles are given with "thetha" parameter.
|
||||
* rotation: If ``true``, 90 degree rotation will be applied on image.
|
||||
* binarization: If ``true``,Otsu thresholding will be applied to augment the input data with binarized images.
|
||||
* scaling_bluring: If ``true``, combination of scaling and blurring will be applied on image.
|
||||
* scaling_binarization: If ``true``, combination of scaling and binarization will be applied on image.
|
||||
* scaling_flip: If ``true``, combination of scaling and flip will be applied on image.
|
||||
* flip_index: Type of flips.
|
||||
* blur_k: Type of blurrings.
|
||||
* scales: Scales of scaling.
|
||||
* brightness: The amount of brightenings.
|
||||
* thetha: Rotation angles.
|
||||
* degrade_scales: The amount of degradings.
|
||||
* continue_training: If ``true``, it means that you have already trained a model and you would like to continue the training. So it is needed to provide the dir of trained model with "dir_of_start_model" and index for naming the models. For example if you have already trained for 3 epochs then your last index is 2 and if you want to continue from model_1.h5, you can set ``index_start`` to 3 to start naming model with index 3.
|
||||
* weighted_loss: If ``true``, this means that you want to apply weighted categorical_crossentropy as loss fucntion. Be carefull if you set to ``true``the parameter "is_loss_soft_dice" should be ``false``
|
||||
* data_is_provided: If you have already provided the input data you can set this to ``true``. Be sure that the train and eval data are in "dir_output". Since when once we provide training data we resize and augment them and then we write them in sub-directories train and eval in "dir_output".
|
||||
* dir_train: This is the directory of "images" and "labels" (dir_train should include two subdirectories with names of images and labels ) for raw images and labels. Namely they are not prepared (not resized and not augmented) yet for training the model. When we run this tool these raw data will be transformed to suitable size needed for the model and they will be written in "dir_output" in train and eval directories. Each of train and eval include "images" and "labels" sub-directories.
|
||||
* index_start: Starting index for saved models in the case that "continue_training" is ``true``.
|
||||
* dir_of_start_model: Directory containing pretrained model to continue training the model in the case that "continue_training" is ``true``.
|
||||
* transformer_num_patches_xy: Number of patches for vision transformer in x and y direction respectively.
|
||||
* transformer_patchsize_x: Patch size of vision transformer patches in x direction.
|
||||
* transformer_patchsize_y: Patch size of vision transformer patches in y direction.
|
||||
* transformer_projection_dim: Transformer projection dimension. Default value is 64.
|
||||
* transformer_mlp_head_units: Transformer Multilayer Perceptron (MLP) head units. Default value is [128, 64].
|
||||
* transformer_layers: transformer layers. Default value is 8.
|
||||
* transformer_num_heads: Transformer number of heads. Default value is 4.
|
||||
* transformer_cnn_first: We have two types of vision transformers. In one type, a CNN is applied first, followed by a transformer. In the other type, this order is reversed. If transformer_cnn_first is true, it means the CNN will be applied before the transformer. Default value is true.
|
||||
|
||||
In the case of segmentation and enhancement the train and evaluation directory should be as following.
|
||||
|
||||
|
|
@ -432,39 +349,12 @@ And the "dir_eval" the same structure as train directory:
|
|||
└── labels # directory of labels
|
||||
```
|
||||
|
||||
After configuring the JSON file for segmentation or enhancement, training can be initiated by running the following
|
||||
After configuring the JSON file for segmentation or enhancement, training can be initiated by running the following
|
||||
command, similar to the process for classification and reading order:
|
||||
|
||||
```
|
||||
eynollah-training train with config_classification.json`
|
||||
```
|
||||
`python train.py with config_classification.json`
|
||||
|
||||
#### Binarization
|
||||
|
||||
### Ground truth format
|
||||
|
||||
Lables for each pixel are identified by a number. So if you have a
|
||||
binary case, ``n_classes`` should be set to ``2`` and labels should
|
||||
be ``0`` and ``1`` for each class and pixel.
|
||||
|
||||
In the case of multiclass, just set ``n_classes`` to the number of classes
|
||||
you have and the try to produce the labels by pixels set from ``0 , 1 ,2 .., n_classes-1``.
|
||||
The labels format should be png.
|
||||
Our lables are 3 channel png images but only information of first channel is used.
|
||||
If you have an image label with height and width of 10, for a binary case the first channel should look like this:
|
||||
|
||||
Label: [ [1, 0, 0, 1, 1, 0, 0, 1, 0, 0],
|
||||
[0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
|
||||
...,
|
||||
[0, 0, 0, 0, 0, 0, 0, 0, 0, 0],
|
||||
[0, 0, 0, 0, 0, 0, 0, 0, 0, 0] ]
|
||||
|
||||
This means that you have an image by `10*10*3` and `pixel[0,0]` belongs
|
||||
to class `1` and `pixel[0,1]` belongs to class `0`.
|
||||
|
||||
A small sample of training data for binarization experiment can be found here, [Training data sample](https://qurator-data.de/~vahid.rezanezhad/binarization_training_data_sample/), which contains images and lables folders.
|
||||
|
||||
|
||||
An example config json file for binarization can be like this:
|
||||
|
||||
```yaml
|
||||
|
|
@ -508,7 +398,7 @@ An example config json file for binarization can be like this:
|
|||
"thetha" : [10, -10],
|
||||
"continue_training": false,
|
||||
"index_start" : 0,
|
||||
"dir_of_start_model" : " ",
|
||||
"dir_of_start_model" : " ",
|
||||
"weighted_loss": false,
|
||||
"is_loss_soft_dice": false,
|
||||
"data_is_provided": false,
|
||||
|
|
@ -553,7 +443,7 @@ An example config json file for binarization can be like this:
|
|||
"thetha" : [10, -10],
|
||||
"continue_training": false,
|
||||
"index_start" : 0,
|
||||
"dir_of_start_model" : " ",
|
||||
"dir_of_start_model" : " ",
|
||||
"weighted_loss": false,
|
||||
"is_loss_soft_dice": false,
|
||||
"data_is_provided": false,
|
||||
|
|
@ -598,7 +488,7 @@ An example config json file for binarization can be like this:
|
|||
"thetha" : [10, -10],
|
||||
"continue_training": false,
|
||||
"index_start" : 0,
|
||||
"dir_of_start_model" : " ",
|
||||
"dir_of_start_model" : " ",
|
||||
"weighted_loss": false,
|
||||
"is_loss_soft_dice": false,
|
||||
"data_is_provided": false,
|
||||
|
|
@ -608,7 +498,7 @@ An example config json file for binarization can be like this:
|
|||
}
|
||||
```
|
||||
|
||||
It's important to mention that the value of n_classes for enhancement should be 3, as the model's output is a 3-channel
|
||||
It's important to mention that the value of n_classes for enhancement should be 3, as the model's output is a 3-channel
|
||||
image.
|
||||
|
||||
#### Page extraction
|
||||
|
|
@ -646,7 +536,7 @@ image.
|
|||
"thetha" : [10, -10],
|
||||
"continue_training": false,
|
||||
"index_start" : 0,
|
||||
"dir_of_start_model" : " ",
|
||||
"dir_of_start_model" : " ",
|
||||
"weighted_loss": false,
|
||||
"is_loss_soft_dice": false,
|
||||
"data_is_provided": false,
|
||||
|
|
@ -656,11 +546,10 @@ image.
|
|||
}
|
||||
```
|
||||
|
||||
For page segmentation (or print space or border segmentation), the model needs to view the input image in its
|
||||
entirety,hence the patches parameter should be set to false.
|
||||
For page segmentation (or printspace or border segmentation), the model needs to view the input image in its entirety,
|
||||
hence the patches parameter should be set to false.
|
||||
|
||||
#### layout segmentation
|
||||
|
||||
An example config json file for layout segmentation with 5 classes (including background) can be like this:
|
||||
|
||||
```yaml
|
||||
|
|
@ -704,7 +593,7 @@ An example config json file for layout segmentation with 5 classes (including ba
|
|||
"thetha" : [10, -10],
|
||||
"continue_training": false,
|
||||
"index_start" : 0,
|
||||
"dir_of_start_model" : " ",
|
||||
"dir_of_start_model" : " ",
|
||||
"weighted_loss": false,
|
||||
"is_loss_soft_dice": false,
|
||||
"data_is_provided": false,
|
||||
|
|
@ -716,42 +605,28 @@ An example config json file for layout segmentation with 5 classes (including ba
|
|||
## Inference with the trained model
|
||||
|
||||
### classification
|
||||
|
||||
For conducting inference with a trained model, you simply need to execute the following command line, specifying the
|
||||
For conducting inference with a trained model, you simply need to execute the following command line, specifying the
|
||||
directory of the model and the image on which to perform inference:
|
||||
|
||||
```sh
|
||||
eynollah-training inference -m "model dir" -i "image"
|
||||
```
|
||||
`python inference.py -m "model dir" -i "image" `
|
||||
|
||||
This will straightforwardly return the class of the image.
|
||||
|
||||
### machine based reading order
|
||||
To infer the reading order using a reading order model, we need a page XML file containing layout information but
|
||||
without the reading order. We simply need to provide the model directory, the XML file, and the output directory.
|
||||
The new XML file with the added reading order will be written to the output directory with the same name.
|
||||
We need to run:
|
||||
|
||||
To infer the reading order using a reading order model, we need a page XML file containing layout information but
|
||||
without the reading order. We simply need to provide the model directory, the XML file, and the output directory. The
|
||||
new XML file with the added reading order will be written to the output directory with the same name. We need to run:
|
||||
|
||||
```sh
|
||||
eynollah-training inference \
|
||||
-m "model dir" \
|
||||
-xml "page xml file" \
|
||||
-o "output dir to write new xml with reading order"
|
||||
```
|
||||
`python inference.py -m "model dir" -xml "page xml file" -o "output dir to write new xml with reading order" `
|
||||
|
||||
### Segmentation (Textline, Binarization, Page extraction and layout) and enhancement
|
||||
For conducting inference with a trained model for segmentation and enhancement you need to run the following command
|
||||
line:
|
||||
|
||||
For conducting inference with a trained model for segmentation and enhancement you need to run the following command line:
|
||||
|
||||
```sh
|
||||
eynollah-training inference \
|
||||
-m "model dir" \
|
||||
-i "image" \
|
||||
-p \
|
||||
-s "output image"
|
||||
```
|
||||
`python inference.py -m "model dir" -i "image" -p -s "output image" `
|
||||
|
||||
Note that in the case of page extraction the -p flag is not needed.
|
||||
|
||||
For segmentation or binarization tasks, if a ground truth (GT) label is available, the IoU evaluation metric can be
|
||||
For segmentation or binarization tasks, if a ground truth (GT) label is available, the IoU evaluation metric can be
|
||||
calculated for the output. To do this, you need to provide the GT label using the argument -gt.
|
||||
|
|
|
|||
|
|
@ -11,18 +11,9 @@ description = "Document Layout Analysis"
|
|||
readme = "README.md"
|
||||
license.file = "LICENSE"
|
||||
requires-python = ">=3.8"
|
||||
keywords = [
|
||||
"document layout analysis",
|
||||
"image segmentation",
|
||||
"binarization",
|
||||
"optical character recognition"
|
||||
]
|
||||
keywords = ["document layout analysis", "image segmentation"]
|
||||
|
||||
dynamic = [
|
||||
"dependencies",
|
||||
"optional-dependencies",
|
||||
"version"
|
||||
]
|
||||
dynamic = ["dependencies", "version"]
|
||||
|
||||
classifiers = [
|
||||
"Development Status :: 4 - Beta",
|
||||
|
|
@ -30,17 +21,16 @@ classifiers = [
|
|||
"Intended Audience :: Science/Research",
|
||||
"License :: OSI Approved :: Apache Software License",
|
||||
"Programming Language :: Python :: 3",
|
||||
"Programming Language :: Python :: 3.8",
|
||||
"Programming Language :: Python :: 3.9",
|
||||
"Programming Language :: Python :: 3.10",
|
||||
"Programming Language :: Python :: 3.11",
|
||||
"Programming Language :: Python :: 3 :: Only",
|
||||
"Topic :: Scientific/Engineering :: Image Processing",
|
||||
]
|
||||
|
||||
[project.optional-dependencies]
|
||||
OCR = ["torch <= 2.0.1", "transformers <= 4.30.2"]
|
||||
plotting = ["matplotlib"]
|
||||
|
||||
[project.scripts]
|
||||
eynollah = "eynollah.cli:main"
|
||||
eynollah-training = "eynollah.training.cli:main"
|
||||
ocrd-eynollah-segment = "eynollah.ocrd_cli:main"
|
||||
ocrd-sbb-binarize = "eynollah.ocrd_cli_binarization:main"
|
||||
|
||||
|
|
@ -51,35 +41,13 @@ Repository = "https://github.com/qurator-spk/eynollah.git"
|
|||
[tool.setuptools.dynamic]
|
||||
dependencies = {file = ["requirements.txt"]}
|
||||
optional-dependencies.test = {file = ["requirements-test.txt"]}
|
||||
optional-dependencies.OCR = {file = ["requirements-ocr.txt"]}
|
||||
optional-dependencies.plotting = {file = ["requirements-plotting.txt"]}
|
||||
optional-dependencies.training = {file = ["requirements-training.txt"]}
|
||||
|
||||
[tool.setuptools.packages.find]
|
||||
where = ["src"]
|
||||
|
||||
[tool.setuptools.package-data]
|
||||
"*" = ["*.json", '*.yml', '*.xml', '*.xsd', '*.ttf']
|
||||
"*" = ["*.json", '*.yml', '*.xml', '*.xsd']
|
||||
|
||||
[tool.coverage.run]
|
||||
branch = true
|
||||
source = ["eynollah"]
|
||||
|
||||
[tool.ruff]
|
||||
line-length = 120
|
||||
|
||||
[tool.ruff.lint]
|
||||
ignore = [
|
||||
# disable unused imports
|
||||
"F401",
|
||||
# disable import order
|
||||
"E402",
|
||||
# disable unused variables
|
||||
"F841",
|
||||
# disable bare except
|
||||
"E722",
|
||||
]
|
||||
|
||||
[tool.ruff.format]
|
||||
quote-style = "preserve"
|
||||
|
||||
|
|
|
|||
|
|
@ -1,2 +0,0 @@
|
|||
torch
|
||||
transformers <= 4.30.2
|
||||
|
|
@ -1 +0,0 @@
|
|||
matplotlib
|
||||
|
|
@ -1,4 +1,4 @@
|
|||
pytest
|
||||
pytest-isolate
|
||||
pytest-subtests
|
||||
coverage[toml]
|
||||
black
|
||||
|
|
|
|||
|
|
@ -1 +0,0 @@
|
|||
train/requirements.txt
|
||||
|
|
@ -4,6 +4,4 @@ numpy <1.24.0
|
|||
scikit-learn >= 0.23.2
|
||||
tensorflow < 2.13
|
||||
numba <= 0.58.1
|
||||
scikit-image
|
||||
biopython
|
||||
tabulate
|
||||
loky
|
||||
|
|
|
|||
Binary file not shown.
404
src/eynollah/cli.py
Normal file
404
src/eynollah/cli.py
Normal file
|
|
@ -0,0 +1,404 @@
|
|||
import sys
|
||||
import click
|
||||
from ocrd_utils import initLogging, getLevelName, getLogger
|
||||
from eynollah.eynollah import Eynollah, Eynollah_ocr
|
||||
from eynollah.sbb_binarize import SbbBinarizer
|
||||
|
||||
@click.group()
|
||||
def main():
|
||||
pass
|
||||
|
||||
@main.command()
|
||||
@click.option(
|
||||
"--dir_xml",
|
||||
"-dx",
|
||||
help="directory of GT page-xml files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_out_modal_image",
|
||||
"-domi",
|
||||
help="directory where ground truth images would be written",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_out_classes",
|
||||
"-docl",
|
||||
help="directory where ground truth classes would be written",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--input_height",
|
||||
"-ih",
|
||||
help="input height",
|
||||
)
|
||||
@click.option(
|
||||
"--input_width",
|
||||
"-iw",
|
||||
help="input width",
|
||||
)
|
||||
@click.option(
|
||||
"--min_area_size",
|
||||
"-min",
|
||||
help="min area size of regions considered for reading order training.",
|
||||
)
|
||||
def machine_based_reading_order(dir_xml, dir_out_modal_image, dir_out_classes, input_height, input_width, min_area_size):
|
||||
xml_files_ind = os.listdir(dir_xml)
|
||||
|
||||
@main.command()
|
||||
@click.option('--patches/--no-patches', default=True, help='by enabling this parameter you let the model to see the image in patches.')
|
||||
@click.option('--model_dir', '-m', type=click.Path(exists=True, file_okay=False), required=True, help='directory containing models for prediction')
|
||||
@click.argument('input_image', required=False)
|
||||
@click.argument('output_image', required=False)
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of input images",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_out",
|
||||
"-do",
|
||||
help="directory for output images",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
def binarization(patches, model_dir, input_image, output_image, dir_in, dir_out):
|
||||
assert (dir_out is None) == (dir_in is None), "Options -di and -do are mutually dependent"
|
||||
assert (input_image is None) == (output_image is None), "INPUT_IMAGE and OUTPUT_IMAGE are mutually dependent"
|
||||
assert (dir_in is None) != (input_image is None), "Specify either -di and -do options, or INPUT_IMAGE and OUTPUT_IMAGE"
|
||||
SbbBinarizer(model_dir).run(image_path=input_image, use_patches=patches, save=output_image, dir_in=dir_in, dir_out=dir_out)
|
||||
|
||||
|
||||
|
||||
|
||||
@main.command()
|
||||
@click.option(
|
||||
"--image",
|
||||
"-i",
|
||||
help="image filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--out",
|
||||
"-o",
|
||||
help="directory to write output xml data",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--overwrite",
|
||||
"-O",
|
||||
help="overwrite (instead of skipping) if output xml exists",
|
||||
is_flag=True,
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of images",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--model",
|
||||
"-m",
|
||||
help="directory of models",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--save_images",
|
||||
"-si",
|
||||
help="if a directory is given, images in documents will be cropped and saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--save_layout",
|
||||
"-sl",
|
||||
help="if a directory is given, plot of layout will be saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--save_deskewed",
|
||||
"-sd",
|
||||
help="if a directory is given, deskewed image will be saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--save_all",
|
||||
"-sa",
|
||||
help="if a directory is given, all plots needed for documentation will be saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--save_page",
|
||||
"-sp",
|
||||
help="if a directory is given, page crop of image will be saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--enable-plotting/--disable-plotting",
|
||||
"-ep/-noep",
|
||||
is_flag=True,
|
||||
help="If set, will plot intermediary files and images",
|
||||
)
|
||||
@click.option(
|
||||
"--extract_only_images/--disable-extracting_only_images",
|
||||
"-eoi/-noeoi",
|
||||
is_flag=True,
|
||||
help="If a directory is given, only images in documents will be cropped and saved there and the other processing will not be done",
|
||||
)
|
||||
@click.option(
|
||||
"--allow-enhancement/--no-allow-enhancement",
|
||||
"-ae/-noae",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would check that input image need resizing and enhancement or not. If so output of resized and enhanced image and corresponding layout data will be written in out directory",
|
||||
)
|
||||
@click.option(
|
||||
"--curved-line/--no-curvedline",
|
||||
"-cl/-nocl",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will try to return contoure of textlines instead of rectangle bounding box of textline. This should be taken into account that with this option the tool need more time to do process.",
|
||||
)
|
||||
@click.option(
|
||||
"--textline_light/--no-textline_light",
|
||||
"-tll/-notll",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will try to return contoure of textlines instead of rectangle bounding box of textline with a faster method.",
|
||||
)
|
||||
@click.option(
|
||||
"--full-layout/--no-full-layout",
|
||||
"-fl/-nofl",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will try to return all elements of layout.",
|
||||
)
|
||||
@click.option(
|
||||
"--tables/--no-tables",
|
||||
"-tab/-notab",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will try to detect tables.",
|
||||
)
|
||||
@click.option(
|
||||
"--right2left/--left2right",
|
||||
"-r2l/-l2r",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will extract right-to-left reading order.",
|
||||
)
|
||||
@click.option(
|
||||
"--input_binary/--input-RGB",
|
||||
"-ib/-irgb",
|
||||
is_flag=True,
|
||||
help="in general, eynollah uses RGB as input but if the input document is strongly dark, bright or for any other reason you can turn binarized input on. This option does not mean that you have to provide a binary image, otherwise this means that the tool itself will binarized the RGB input document.",
|
||||
)
|
||||
@click.option(
|
||||
"--allow_scaling/--no-allow-scaling",
|
||||
"-as/-noas",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would check the scale and if needed it will scale it to perform better layout detection",
|
||||
)
|
||||
@click.option(
|
||||
"--headers_off/--headers-on",
|
||||
"-ho/-noho",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would ignore headers role in reading order",
|
||||
)
|
||||
@click.option(
|
||||
"--light_version/--original",
|
||||
"-light/-org",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would use lighter version",
|
||||
)
|
||||
@click.option(
|
||||
"--ignore_page_extraction/--extract_page_included",
|
||||
"-ipe/-epi",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would ignore page extraction",
|
||||
)
|
||||
@click.option(
|
||||
"--reading_order_machine_based/--heuristic_reading_order",
|
||||
"-romb/-hro",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would apply machine based reading order detection",
|
||||
)
|
||||
@click.option(
|
||||
"--do_ocr",
|
||||
"-ocr/-noocr",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will try to do ocr",
|
||||
)
|
||||
@click.option(
|
||||
"--num_col_upper",
|
||||
"-ncu",
|
||||
help="lower limit of columns in document image",
|
||||
)
|
||||
@click.option(
|
||||
"--num_col_lower",
|
||||
"-ncl",
|
||||
help="upper limit of columns in document image",
|
||||
)
|
||||
@click.option(
|
||||
"--skip_layout_and_reading_order",
|
||||
"-slro/-noslro",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will ignore layout detection and reading order. It means that textline detection will be done within printspace and contours of textline will be written in xml output file.",
|
||||
)
|
||||
@click.option(
|
||||
"--log_level",
|
||||
"-l",
|
||||
type=click.Choice(['OFF', 'DEBUG', 'INFO', 'WARN', 'ERROR']),
|
||||
help="Override log level globally to this",
|
||||
)
|
||||
|
||||
def layout(image, out, overwrite, dir_in, model, save_images, save_layout, save_deskewed, save_all, extract_only_images, save_page, enable_plotting, allow_enhancement, curved_line, textline_light, full_layout, tables, right2left, input_binary, allow_scaling, headers_off, light_version, reading_order_machine_based, do_ocr, num_col_upper, num_col_lower, skip_layout_and_reading_order, ignore_page_extraction, log_level):
|
||||
initLogging()
|
||||
if log_level:
|
||||
getLogger('eynollah').setLevel(getLevelName(log_level))
|
||||
assert enable_plotting or not save_layout, "Plotting with -sl also requires -ep"
|
||||
assert enable_plotting or not save_deskewed, "Plotting with -sd also requires -ep"
|
||||
assert enable_plotting or not save_all, "Plotting with -sa also requires -ep"
|
||||
assert enable_plotting or not save_page, "Plotting with -sp also requires -ep"
|
||||
assert enable_plotting or not save_images, "Plotting with -si also requires -ep"
|
||||
assert enable_plotting or not allow_enhancement, "Plotting with -ae also requires -ep"
|
||||
assert not enable_plotting or save_layout or save_deskewed or save_all or save_page or save_images or allow_enhancement, \
|
||||
"Plotting with -ep also requires -sl, -sd, -sa, -sp, -si or -ae"
|
||||
assert textline_light == light_version, "Both light textline detection -tll and light version -light must be set or unset equally"
|
||||
assert not extract_only_images or not allow_enhancement, "Image extraction -eoi can not be set alongside allow_enhancement -ae"
|
||||
assert not extract_only_images or not allow_scaling, "Image extraction -eoi can not be set alongside allow_scaling -as"
|
||||
assert not extract_only_images or not light_version, "Image extraction -eoi can not be set alongside light_version -light"
|
||||
assert not extract_only_images or not curved_line, "Image extraction -eoi can not be set alongside curved_line -cl"
|
||||
assert not extract_only_images or not textline_light, "Image extraction -eoi can not be set alongside textline_light -tll"
|
||||
assert not extract_only_images or not full_layout, "Image extraction -eoi can not be set alongside full_layout -fl"
|
||||
assert not extract_only_images or not tables, "Image extraction -eoi can not be set alongside tables -tab"
|
||||
assert not extract_only_images or not right2left, "Image extraction -eoi can not be set alongside right2left -r2l"
|
||||
assert not extract_only_images or not headers_off, "Image extraction -eoi can not be set alongside headers_off -ho"
|
||||
assert image or dir_in, "Either a single image -i or a dir_in -di is required"
|
||||
eynollah = Eynollah(
|
||||
model,
|
||||
logger=getLogger('eynollah'),
|
||||
dir_out=out,
|
||||
dir_of_cropped_images=save_images,
|
||||
extract_only_images=extract_only_images,
|
||||
dir_of_layout=save_layout,
|
||||
dir_of_deskewed=save_deskewed,
|
||||
dir_of_all=save_all,
|
||||
dir_save_page=save_page,
|
||||
enable_plotting=enable_plotting,
|
||||
allow_enhancement=allow_enhancement,
|
||||
curved_line=curved_line,
|
||||
textline_light=textline_light,
|
||||
full_layout=full_layout,
|
||||
tables=tables,
|
||||
right2left=right2left,
|
||||
input_binary=input_binary,
|
||||
allow_scaling=allow_scaling,
|
||||
headers_off=headers_off,
|
||||
light_version=light_version,
|
||||
ignore_page_extraction=ignore_page_extraction,
|
||||
reading_order_machine_based=reading_order_machine_based,
|
||||
do_ocr=do_ocr,
|
||||
num_col_upper=num_col_upper,
|
||||
num_col_lower=num_col_lower,
|
||||
skip_layout_and_reading_order=skip_layout_and_reading_order,
|
||||
)
|
||||
if dir_in:
|
||||
eynollah.run(dir_in=dir_in, overwrite=overwrite)
|
||||
else:
|
||||
eynollah.run(image_filename=image, overwrite=overwrite)
|
||||
|
||||
|
||||
@main.command()
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of images",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in_bin",
|
||||
"-dib",
|
||||
help="directory of binarized images. This should be given if you want to do prediction based on both rgb and bin images. And all bin images are png files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--out",
|
||||
"-o",
|
||||
help="directory to write output xml data",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--dir_xmls",
|
||||
"-dx",
|
||||
help="directory of xmls",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_out_image_text",
|
||||
"-doit",
|
||||
help="directory of images with predicted text",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--model",
|
||||
"-m",
|
||||
help="directory of models",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--tr_ocr",
|
||||
"-trocr/-notrocr",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, transformer ocr will be applied, otherwise cnn_rnn model.",
|
||||
)
|
||||
@click.option(
|
||||
"--export_textline_images_and_text",
|
||||
"-etit/-noetit",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, images and text in xml will be exported into output dir. This files can be used for training a OCR engine.",
|
||||
)
|
||||
@click.option(
|
||||
"--do_not_mask_with_textline_contour",
|
||||
"-nmtc/-mtc",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, cropped textline images will not be masked with textline contour.",
|
||||
)
|
||||
@click.option(
|
||||
"--draw_texts_on_image",
|
||||
"-dtoi/-ndtoi",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, the predicted texts will be displayed on an image.",
|
||||
)
|
||||
@click.option(
|
||||
"--prediction_with_both_of_rgb_and_bin",
|
||||
"-brb/-nbrb",
|
||||
is_flag=True,
|
||||
help="If this parameter is set to True, the prediction will be performed using both RGB and binary images. However, this does not necessarily improve results; it may be beneficial for certain document images.",
|
||||
)
|
||||
@click.option(
|
||||
"--log_level",
|
||||
"-l",
|
||||
type=click.Choice(['OFF', 'DEBUG', 'INFO', 'WARN', 'ERROR']),
|
||||
help="Override log level globally to this",
|
||||
)
|
||||
|
||||
def ocr(dir_in, dir_in_bin, out, dir_xmls, dir_out_image_text, model, tr_ocr, export_textline_images_and_text, do_not_mask_with_textline_contour, draw_texts_on_image, prediction_with_both_of_rgb_and_bin, log_level):
|
||||
initLogging()
|
||||
if log_level:
|
||||
getLogger('eynollah').setLevel(getLevelName(log_level))
|
||||
eynollah_ocr = Eynollah_ocr(
|
||||
dir_xmls=dir_xmls,
|
||||
dir_out_image_text=dir_out_image_text,
|
||||
dir_in=dir_in,
|
||||
dir_in_bin=dir_in_bin,
|
||||
dir_out=out,
|
||||
dir_models=model,
|
||||
tr_ocr=tr_ocr,
|
||||
export_textline_images_and_text=export_textline_images_and_text,
|
||||
do_not_mask_with_textline_contour=do_not_mask_with_textline_contour,
|
||||
draw_texts_on_image=draw_texts_on_image,
|
||||
prediction_with_both_of_rgb_and_bin=prediction_with_both_of_rgb_and_bin,
|
||||
)
|
||||
eynollah_ocr.run()
|
||||
|
||||
if __name__ == "__main__":
|
||||
main()
|
||||
|
|
@ -1,22 +0,0 @@
|
|||
# NOTE: For predictable order of imports of torch/shapely/tensorflow
|
||||
# this must be the first import of the CLI!
|
||||
from ..eynollah_imports import imported_libs
|
||||
|
||||
from .cli_models import models_cli
|
||||
from .cli_binarize import binarize_cli
|
||||
|
||||
from .cli import main
|
||||
from .cli_binarize import binarize_cli
|
||||
from .cli_enhance import enhance_cli
|
||||
from .cli_extract_images import extract_images_cli
|
||||
from .cli_layout import layout_cli
|
||||
from .cli_ocr import ocr_cli
|
||||
from .cli_readingorder import readingorder_cli
|
||||
|
||||
main.add_command(binarize_cli, 'binarization')
|
||||
main.add_command(enhance_cli, 'enhancement')
|
||||
main.add_command(layout_cli, 'layout')
|
||||
main.add_command(readingorder_cli, 'machine-based-reading-order')
|
||||
main.add_command(models_cli, 'models')
|
||||
main.add_command(ocr_cli, 'ocr')
|
||||
main.add_command(extract_images_cli, 'extract-images')
|
||||
|
|
@ -1,66 +0,0 @@
|
|||
from dataclasses import dataclass
|
||||
import logging
|
||||
import sys
|
||||
import os
|
||||
from typing import Union
|
||||
|
||||
import click
|
||||
|
||||
from ..model_zoo import EynollahModelZoo
|
||||
from .cli_models import models_cli
|
||||
|
||||
@dataclass()
|
||||
class EynollahCliCtx:
|
||||
"""
|
||||
Holds options relevant for all eynollah subcommands
|
||||
"""
|
||||
model_zoo: EynollahModelZoo
|
||||
log_level : Union[str, None] = 'INFO'
|
||||
|
||||
|
||||
@click.group()
|
||||
@click.option(
|
||||
"--model-basedir",
|
||||
"-m",
|
||||
help="directory of models",
|
||||
# NOTE: not mandatory to exist so --help for subcommands works but will log a warning
|
||||
# and raise exception when trying to load models in the CLI
|
||||
# type=click.Path(exists=True),
|
||||
default=f'{os.getcwd()}/models_eynollah',
|
||||
)
|
||||
@click.option(
|
||||
"--model-overrides",
|
||||
"-mv",
|
||||
help="override default versions of model categories, syntax is 'CATEGORY VARIANT PATH', e.g 'region light /path/to/model'. See eynollah list-models for the full list",
|
||||
type=(str, str, str),
|
||||
multiple=True,
|
||||
)
|
||||
@click.option(
|
||||
"--log_level",
|
||||
"-l",
|
||||
type=click.Choice(['OFF', 'DEBUG', 'INFO', 'WARN', 'ERROR']),
|
||||
help="Override log level globally to this",
|
||||
)
|
||||
@click.pass_context
|
||||
def main(ctx, model_basedir, model_overrides, log_level):
|
||||
"""
|
||||
eynollah - Document Layout Analysis, Image Enhancement, OCR
|
||||
"""
|
||||
# Initialize logging
|
||||
console_handler = logging.StreamHandler(sys.stderr)
|
||||
console_handler.setLevel(logging.NOTSET)
|
||||
formatter = logging.Formatter('%(asctime)s.%(msecs)03d %(levelname)s %(name)s - %(message)s', datefmt='%H:%M:%S')
|
||||
console_handler.setFormatter(formatter)
|
||||
logging.getLogger('eynollah').addHandler(console_handler)
|
||||
logging.getLogger('eynollah').setLevel(log_level or logging.INFO)
|
||||
# Initialize model zoo
|
||||
model_zoo = EynollahModelZoo(basedir=model_basedir, model_overrides=model_overrides)
|
||||
# Initialize CLI context
|
||||
ctx.obj = EynollahCliCtx(
|
||||
model_zoo=model_zoo,
|
||||
log_level=log_level,
|
||||
)
|
||||
|
||||
|
||||
if __name__ == "__main__":
|
||||
main()
|
||||
|
|
@ -1,44 +0,0 @@
|
|||
import click
|
||||
|
||||
@click.command()
|
||||
@click.option('--patches/--no-patches', default=True, help='by enabling this parameter you let the model to see the image in patches.')
|
||||
@click.option(
|
||||
"--input-image", "--image",
|
||||
"-i",
|
||||
help="input image filename",
|
||||
type=click.Path(exists=True, dir_okay=False)
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of input images (instead of --image)",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--output",
|
||||
"-o",
|
||||
help="output image (if using -i) or output image directory (if using -di)",
|
||||
type=click.Path(file_okay=True, dir_okay=True),
|
||||
required=True,
|
||||
)
|
||||
@click.pass_context
|
||||
def binarize_cli(
|
||||
ctx,
|
||||
patches,
|
||||
input_image,
|
||||
dir_in,
|
||||
output,
|
||||
):
|
||||
"""
|
||||
Binarize images with a ML model
|
||||
"""
|
||||
from ..sbb_binarize import SbbBinarizer
|
||||
assert bool(input_image) != bool(dir_in), "Either -i (single input) or -di (directory) must be provided, but not both."
|
||||
binarizer = SbbBinarizer(model_zoo=ctx.obj.model_zoo)
|
||||
binarizer.run(
|
||||
image_path=input_image,
|
||||
use_patches=patches,
|
||||
output=output,
|
||||
dir_in=dir_in
|
||||
)
|
||||
|
||||
|
|
@ -1,63 +0,0 @@
|
|||
import click
|
||||
|
||||
@click.command()
|
||||
@click.option(
|
||||
"--image",
|
||||
"-i",
|
||||
help="input image filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--out",
|
||||
"-o",
|
||||
help="directory for output PAGE-XML files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--overwrite",
|
||||
"-O",
|
||||
help="overwrite (instead of skipping) if output xml exists",
|
||||
is_flag=True,
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of input images (instead of --image)",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--num_col_upper",
|
||||
"-ncu",
|
||||
help="lower limit of columns in document image",
|
||||
)
|
||||
@click.option(
|
||||
"--num_col_lower",
|
||||
"-ncl",
|
||||
help="upper limit of columns in document image",
|
||||
)
|
||||
@click.option(
|
||||
"--save_org_scale/--no_save_org_scale",
|
||||
"-sos/-nosos",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will save the enhanced image in org scale.",
|
||||
)
|
||||
@click.pass_context
|
||||
def enhance_cli(ctx, image, out, overwrite, dir_in, num_col_upper, num_col_lower, save_org_scale):
|
||||
"""
|
||||
Enhance image
|
||||
"""
|
||||
assert bool(image) != bool(dir_in), "Either -i (single input) or -di (directory) must be provided, but not both."
|
||||
from ..image_enhancer import Enhancer
|
||||
enhancer = Enhancer(
|
||||
model_zoo=ctx.obj.model_zoo,
|
||||
num_col_upper=num_col_upper,
|
||||
num_col_lower=num_col_lower,
|
||||
save_org_scale=save_org_scale,
|
||||
)
|
||||
enhancer.run(overwrite=overwrite,
|
||||
dir_in=dir_in,
|
||||
image_filename=image,
|
||||
dir_out=out,
|
||||
)
|
||||
|
||||
|
|
@ -1,100 +0,0 @@
|
|||
import click
|
||||
|
||||
@click.command()
|
||||
@click.option(
|
||||
"--image",
|
||||
"-i",
|
||||
help="input image filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--out",
|
||||
"-o",
|
||||
help="directory for output PAGE-XML files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--overwrite",
|
||||
"-O",
|
||||
help="overwrite (instead of skipping) if output xml exists",
|
||||
is_flag=True,
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of input images (instead of --image)",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--save_images",
|
||||
"-si",
|
||||
help="if a directory is given, images in documents will be cropped and saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--enable-plotting/--disable-plotting",
|
||||
"-ep/-noep",
|
||||
is_flag=True,
|
||||
help="If set, will plot intermediary files and images",
|
||||
)
|
||||
@click.option(
|
||||
"--input_binary/--input-RGB",
|
||||
"-ib/-irgb",
|
||||
is_flag=True,
|
||||
help="In general, eynollah uses RGB as input but if the input document is very dark, very bright or for any other reason you can turn on input binarization. When this flag is set, eynollah will binarize the RGB input document, you should always provide RGB images to eynollah.",
|
||||
)
|
||||
@click.option(
|
||||
"--ignore_page_extraction/--extract_page_included",
|
||||
"-ipe/-epi",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would ignore page extraction",
|
||||
)
|
||||
@click.option(
|
||||
"--num_col_upper",
|
||||
"-ncu",
|
||||
help="lower limit of columns in document image",
|
||||
)
|
||||
@click.option(
|
||||
"--num_col_lower",
|
||||
"-ncl",
|
||||
help="upper limit of columns in document image",
|
||||
)
|
||||
@click.pass_context
|
||||
def extract_images_cli(
|
||||
ctx,
|
||||
image,
|
||||
out,
|
||||
overwrite,
|
||||
dir_in,
|
||||
save_images,
|
||||
enable_plotting,
|
||||
input_binary,
|
||||
num_col_upper,
|
||||
num_col_lower,
|
||||
ignore_page_extraction,
|
||||
):
|
||||
"""
|
||||
Detect Layout (with optional image enhancement and reading order detection)
|
||||
"""
|
||||
assert enable_plotting or not save_images, "Plotting with -si also requires -ep"
|
||||
assert not enable_plotting or save_images, "Plotting with -ep also requires -si"
|
||||
assert bool(image) != bool(dir_in), "Either -i (single input) or -di (directory) must be provided, but not both."
|
||||
|
||||
from ..extract_images import EynollahImageExtractor
|
||||
extractor = EynollahImageExtractor(
|
||||
model_zoo=ctx.obj.model_zoo,
|
||||
enable_plotting=enable_plotting,
|
||||
input_binary=input_binary,
|
||||
ignore_page_extraction=ignore_page_extraction,
|
||||
num_col_upper=num_col_upper,
|
||||
num_col_lower=num_col_lower,
|
||||
)
|
||||
extractor.run(overwrite=overwrite,
|
||||
image_filename=image,
|
||||
dir_in=dir_in,
|
||||
dir_out=out,
|
||||
dir_of_cropped_images=save_images,
|
||||
)
|
||||
|
||||
|
|
@ -1,223 +0,0 @@
|
|||
import click
|
||||
|
||||
@click.command()
|
||||
@click.option(
|
||||
"--image",
|
||||
"-i",
|
||||
help="input image filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--out",
|
||||
"-o",
|
||||
help="directory for output PAGE-XML files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--overwrite",
|
||||
"-O",
|
||||
help="overwrite (instead of skipping) if output xml exists",
|
||||
is_flag=True,
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of input images (instead of --image)",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--save_images",
|
||||
"-si",
|
||||
help="if a directory is given, images in documents will be cropped and saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--save_layout",
|
||||
"-sl",
|
||||
help="if a directory is given, plot of layout will be saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--save_deskewed",
|
||||
"-sd",
|
||||
help="if a directory is given, deskewed image will be saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--save_all",
|
||||
"-sa",
|
||||
help="if a directory is given, all plots needed for documentation will be saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--save_page",
|
||||
"-sp",
|
||||
help="if a directory is given, page crop of image will be saved there",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--enable-plotting/--disable-plotting",
|
||||
"-ep/-noep",
|
||||
is_flag=True,
|
||||
help="If set, will plot intermediary files and images",
|
||||
)
|
||||
@click.option(
|
||||
"--allow-enhancement/--no-allow-enhancement",
|
||||
"-ae/-noae",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would check that input image need resizing and enhancement or not. If so output of resized and enhanced image and corresponding layout data will be written in out directory",
|
||||
)
|
||||
@click.option(
|
||||
"--curved-line/--no-curvedline",
|
||||
"-cl/-nocl",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will try to return contoure of textlines instead of rectangle bounding box of textline. This should be taken into account that with this option the tool need more time to do process.",
|
||||
)
|
||||
@click.option(
|
||||
"--full-layout/--no-full-layout",
|
||||
"-fl/-nofl",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will try to return all elements of layout.",
|
||||
)
|
||||
@click.option(
|
||||
"--tables/--no-tables",
|
||||
"-tab/-notab",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will try to detect tables.",
|
||||
)
|
||||
@click.option(
|
||||
"--right2left/--left2right",
|
||||
"-r2l/-l2r",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will extract right-to-left reading order.",
|
||||
)
|
||||
@click.option(
|
||||
"--input_binary/--input-RGB",
|
||||
"-ib/-irgb",
|
||||
is_flag=True,
|
||||
help="In general, eynollah uses RGB as input but if the input document is very dark, very bright or for any other reason you can turn on input binarization. When this flag is set, eynollah will binarize the RGB input document, you should always provide RGB images to eynollah.",
|
||||
)
|
||||
@click.option(
|
||||
"--allow_scaling/--no-allow-scaling",
|
||||
"-as/-noas",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would check the scale and if needed it will scale it to perform better layout detection",
|
||||
)
|
||||
@click.option(
|
||||
"--headers_off/--headers-on",
|
||||
"-ho/-noho",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would ignore headers role in reading order",
|
||||
)
|
||||
@click.option(
|
||||
"--ignore_page_extraction/--extract_page_included",
|
||||
"-ipe/-epi",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would ignore page extraction",
|
||||
)
|
||||
@click.option(
|
||||
"--reading_order_machine_based/--heuristic_reading_order",
|
||||
"-romb/-hro",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool would apply machine based reading order detection",
|
||||
)
|
||||
@click.option(
|
||||
"--num_col_upper",
|
||||
"-ncu",
|
||||
help="lower limit of columns in document image",
|
||||
)
|
||||
@click.option(
|
||||
"--num_col_lower",
|
||||
"-ncl",
|
||||
help="upper limit of columns in document image",
|
||||
)
|
||||
@click.option(
|
||||
"--threshold_art_class_layout",
|
||||
"-tharl",
|
||||
help="threshold of artifical class in the case of layout detection. The default value is 0.1",
|
||||
)
|
||||
@click.option(
|
||||
"--threshold_art_class_textline",
|
||||
"-thart",
|
||||
help="threshold of artifical class in the case of textline detection. The default value is 0.1",
|
||||
)
|
||||
@click.option(
|
||||
"--skip_layout_and_reading_order",
|
||||
"-slro/-noslro",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will ignore layout detection and reading order. It means that textline detection will be done within printspace and contours of textline will be written in xml output file.",
|
||||
)
|
||||
@click.pass_context
|
||||
def layout_cli(
|
||||
ctx,
|
||||
image,
|
||||
out,
|
||||
overwrite,
|
||||
dir_in,
|
||||
save_images,
|
||||
save_layout,
|
||||
save_deskewed,
|
||||
save_all,
|
||||
save_page,
|
||||
enable_plotting,
|
||||
allow_enhancement,
|
||||
curved_line,
|
||||
full_layout,
|
||||
tables,
|
||||
right2left,
|
||||
input_binary,
|
||||
allow_scaling,
|
||||
headers_off,
|
||||
reading_order_machine_based,
|
||||
num_col_upper,
|
||||
num_col_lower,
|
||||
threshold_art_class_textline,
|
||||
threshold_art_class_layout,
|
||||
skip_layout_and_reading_order,
|
||||
ignore_page_extraction,
|
||||
):
|
||||
"""
|
||||
Detect Layout (with optional image enhancement and reading order detection)
|
||||
"""
|
||||
from ..eynollah import Eynollah
|
||||
assert enable_plotting or not save_layout, "Plotting with -sl also requires -ep"
|
||||
assert enable_plotting or not save_deskewed, "Plotting with -sd also requires -ep"
|
||||
assert enable_plotting or not save_all, "Plotting with -sa also requires -ep"
|
||||
assert enable_plotting or not save_page, "Plotting with -sp also requires -ep"
|
||||
assert enable_plotting or not save_images, "Plotting with -si also requires -ep"
|
||||
assert enable_plotting or not allow_enhancement, "Plotting with -ae also requires -ep"
|
||||
assert not enable_plotting or save_layout or save_deskewed or save_all or save_page or save_images or allow_enhancement, \
|
||||
"Plotting with -ep also requires -sl, -sd, -sa, -sp, -si or -ae"
|
||||
assert bool(image) != bool(dir_in), "Either -i (single input) or -di (directory) must be provided, but not both."
|
||||
eynollah = Eynollah(
|
||||
model_zoo=ctx.obj.model_zoo,
|
||||
enable_plotting=enable_plotting,
|
||||
allow_enhancement=allow_enhancement,
|
||||
curved_line=curved_line,
|
||||
full_layout=full_layout,
|
||||
tables=tables,
|
||||
right2left=right2left,
|
||||
input_binary=input_binary,
|
||||
allow_scaling=allow_scaling,
|
||||
headers_off=headers_off,
|
||||
ignore_page_extraction=ignore_page_extraction,
|
||||
reading_order_machine_based=reading_order_machine_based,
|
||||
num_col_upper=num_col_upper,
|
||||
num_col_lower=num_col_lower,
|
||||
skip_layout_and_reading_order=skip_layout_and_reading_order,
|
||||
threshold_art_class_textline=threshold_art_class_textline,
|
||||
threshold_art_class_layout=threshold_art_class_layout,
|
||||
)
|
||||
eynollah.run(overwrite=overwrite,
|
||||
image_filename=image,
|
||||
dir_in=dir_in,
|
||||
dir_out=out,
|
||||
dir_of_cropped_images=save_images,
|
||||
dir_of_layout=save_layout,
|
||||
dir_of_deskewed=save_deskewed,
|
||||
dir_of_all=save_all,
|
||||
dir_save_page=save_page,
|
||||
)
|
||||
|
||||
|
|
@ -1,69 +0,0 @@
|
|||
from pathlib import Path
|
||||
from typing import Set, Tuple
|
||||
import click
|
||||
|
||||
from eynollah.model_zoo.default_specs import MODELS_VERSION
|
||||
|
||||
@click.group()
|
||||
@click.pass_context
|
||||
def models_cli(
|
||||
ctx,
|
||||
):
|
||||
"""
|
||||
Organize models for the various runners in eynollah.
|
||||
"""
|
||||
assert ctx.obj.model_zoo
|
||||
|
||||
|
||||
@models_cli.command('list')
|
||||
@click.pass_context
|
||||
def list_models(
|
||||
ctx,
|
||||
):
|
||||
"""
|
||||
List all the models in the zoo
|
||||
"""
|
||||
print(f"Model basedir: {ctx.obj.model_zoo.model_basedir}")
|
||||
print(f"Model overrides: {ctx.obj.model_zoo.model_overrides}")
|
||||
print(ctx.obj.model_zoo)
|
||||
|
||||
|
||||
@models_cli.command('package')
|
||||
@click.option(
|
||||
'--set-version', '-V', 'version', help="Version to use for packaging", default=MODELS_VERSION, show_default=True
|
||||
)
|
||||
@click.argument('output_dir')
|
||||
@click.pass_context
|
||||
def package(
|
||||
ctx,
|
||||
version,
|
||||
output_dir,
|
||||
):
|
||||
"""
|
||||
Generate shell code to copy all the models in the zoo into properly named folders in OUTPUT_DIR for distribution.
|
||||
|
||||
eynollah models -m SRC package OUTPUT_DIR
|
||||
|
||||
SRC should contain a directory "models_eynollah" containing all the models.
|
||||
"""
|
||||
mkdirs: Set[Path] = set([])
|
||||
copies: Set[Tuple[Path, Path]] = set([])
|
||||
for spec in ctx.obj.model_zoo.specs.specs:
|
||||
# skip these as they are dependent on the ocr model
|
||||
if spec.category in ('num_to_char', 'characters'):
|
||||
continue
|
||||
src: Path = ctx.obj.model_zoo.model_path(spec.category, spec.variant)
|
||||
# Only copy the top-most directory relative to models_eynollah
|
||||
while src.parent.name != 'models_eynollah':
|
||||
src = src.parent
|
||||
for dist in spec.dists:
|
||||
dist_dir = Path(f"{output_dir}/models_{dist}_{version}/models_eynollah")
|
||||
copies.add((src, dist_dir))
|
||||
mkdirs.add(dist_dir)
|
||||
for dir in mkdirs:
|
||||
print(f"mkdir -vp {dir}")
|
||||
for (src, dst) in copies:
|
||||
print(f"cp -vr {src} {dst}")
|
||||
for dir in mkdirs:
|
||||
zip_path = Path(f'../{dir.parent.name}.zip')
|
||||
print(f"(cd {dir}/..; zip -vr {zip_path} models_eynollah)")
|
||||
|
|
@ -1,103 +0,0 @@
|
|||
import click
|
||||
|
||||
@click.command()
|
||||
@click.option(
|
||||
"--image",
|
||||
"-i",
|
||||
help="input image filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of input images (instead of --image)",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in_bin",
|
||||
"-dib",
|
||||
help=("directory of binarized images (in addition to --dir_in for RGB images; filename stems must match the RGB image files, with '.png' \n Perform prediction using both RGB and binary images. (This does not necessarily improve results, however it may be beneficial for certain document images."),
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_xmls",
|
||||
"-dx",
|
||||
help="directory of input PAGE-XML files (in addition to --dir_in; filename stems must match the image files, with '.xml' suffix).",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--out",
|
||||
"-o",
|
||||
help="directory for output PAGE-XML files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--dir_out_image_text",
|
||||
"-doit",
|
||||
help="directory for output images, newly rendered with predicted text",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--overwrite",
|
||||
"-O",
|
||||
help="overwrite (instead of skipping) if output xml exists",
|
||||
is_flag=True,
|
||||
)
|
||||
@click.option(
|
||||
"--tr_ocr",
|
||||
"-trocr/-notrocr",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, transformer ocr will be applied, otherwise cnn_rnn model.",
|
||||
)
|
||||
@click.option(
|
||||
"--do_not_mask_with_textline_contour",
|
||||
"-nmtc/-mtc",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, cropped textline images will not be masked with textline contour.",
|
||||
)
|
||||
@click.option(
|
||||
"--batch_size",
|
||||
"-bs",
|
||||
help="number of inference batch size. Default b_s for trocr and cnn_rnn models are 2 and 8 respectively",
|
||||
)
|
||||
@click.option(
|
||||
"--min_conf_value_of_textline_text",
|
||||
"-min_conf",
|
||||
help="minimum OCR confidence value. Text lines with a confidence value lower than this threshold will not be included in the output XML file.",
|
||||
)
|
||||
@click.pass_context
|
||||
def ocr_cli(
|
||||
ctx,
|
||||
image,
|
||||
dir_in,
|
||||
dir_in_bin,
|
||||
dir_xmls,
|
||||
out,
|
||||
dir_out_image_text,
|
||||
overwrite,
|
||||
tr_ocr,
|
||||
do_not_mask_with_textline_contour,
|
||||
batch_size,
|
||||
min_conf_value_of_textline_text,
|
||||
):
|
||||
"""
|
||||
Recognize text with a CNN/RNN or transformer ML model.
|
||||
"""
|
||||
assert bool(image) ^ bool(dir_in), "Either -i (single image) or -di (directory) must be provided, but not both."
|
||||
from ..eynollah_ocr import Eynollah_ocr
|
||||
eynollah_ocr = Eynollah_ocr(
|
||||
model_zoo=ctx.obj.model_zoo,
|
||||
tr_ocr=tr_ocr,
|
||||
do_not_mask_with_textline_contour=do_not_mask_with_textline_contour,
|
||||
batch_size=batch_size,
|
||||
min_conf_value_of_textline_text=min_conf_value_of_textline_text)
|
||||
eynollah_ocr.run(overwrite=overwrite,
|
||||
dir_in=dir_in,
|
||||
dir_in_bin=dir_in_bin,
|
||||
image_filename=image,
|
||||
dir_xmls=dir_xmls,
|
||||
dir_out_image_text=dir_out_image_text,
|
||||
dir_out=out,
|
||||
)
|
||||
|
|
@ -1,35 +0,0 @@
|
|||
import click
|
||||
|
||||
@click.command()
|
||||
@click.option(
|
||||
"--input",
|
||||
"-i",
|
||||
help="PAGE-XML input filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of PAGE-XML input files (instead of --input)",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--out",
|
||||
"-o",
|
||||
help="directory for output images",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.pass_context
|
||||
def readingorder_cli(ctx, input, dir_in, out):
|
||||
"""
|
||||
Generate ReadingOrder with a ML model
|
||||
"""
|
||||
from ..mb_ro_on_layout import machine_based_reading_order_on_layout
|
||||
assert bool(input) != bool(dir_in), "Either -i (single input) or -di (directory) must be provided, but not both."
|
||||
orderer = machine_based_reading_order_on_layout(model_zoo=ctx.obj.model_zoo)
|
||||
orderer.run(xml_filename=input,
|
||||
dir_in=dir_in,
|
||||
dir_out=out,
|
||||
)
|
||||
|
||||
|
|
@ -1,281 +0,0 @@
|
|||
"""
|
||||
extract images?
|
||||
"""
|
||||
|
||||
from concurrent.futures import ProcessPoolExecutor
|
||||
import logging
|
||||
from multiprocessing import cpu_count
|
||||
import os
|
||||
import time
|
||||
from typing import Optional
|
||||
from pathlib import Path
|
||||
import tensorflow as tf
|
||||
import numpy as np
|
||||
import cv2
|
||||
|
||||
from eynollah.utils.contour import filter_contours_area_of_image, return_contours_of_image, return_contours_of_interested_region
|
||||
from eynollah.utils.resize import resize_image
|
||||
|
||||
from .model_zoo.model_zoo import EynollahModelZoo
|
||||
from .eynollah import Eynollah
|
||||
from .utils import box2rect, is_image_filename
|
||||
from .plot import EynollahPlotter
|
||||
|
||||
class EynollahImageExtractor(Eynollah):
|
||||
|
||||
def __init__(
|
||||
self,
|
||||
*,
|
||||
model_zoo: EynollahModelZoo,
|
||||
enable_plotting : bool = False,
|
||||
input_binary : bool = False,
|
||||
ignore_page_extraction : bool = False,
|
||||
num_col_upper : Optional[int] = None,
|
||||
num_col_lower : Optional[int] = None,
|
||||
full_layout : bool = False,
|
||||
tables : bool = False,
|
||||
curved_line : bool = False,
|
||||
allow_enhancement : bool = False,
|
||||
|
||||
):
|
||||
self.logger = logging.getLogger('eynollah.extract_images')
|
||||
self.model_zoo = model_zoo
|
||||
self.plotter = None
|
||||
self.tables = tables
|
||||
self.curved_line = curved_line
|
||||
self.allow_enhancement = allow_enhancement
|
||||
|
||||
self.enable_plotting = enable_plotting
|
||||
# --input-binary sensible if image is very dark, if layout is not working.
|
||||
self.input_binary = input_binary
|
||||
self.ignore_page_extraction = ignore_page_extraction
|
||||
self.full_layout = full_layout
|
||||
if num_col_upper:
|
||||
self.num_col_upper = int(num_col_upper)
|
||||
else:
|
||||
self.num_col_upper = num_col_upper
|
||||
if num_col_lower:
|
||||
self.num_col_lower = int(num_col_lower)
|
||||
else:
|
||||
self.num_col_lower = num_col_lower
|
||||
|
||||
# for parallelization of CPU-intensive tasks:
|
||||
self.executor = ProcessPoolExecutor(max_workers=cpu_count())
|
||||
|
||||
t_start = time.time()
|
||||
|
||||
try:
|
||||
for device in tf.config.list_physical_devices('GPU'):
|
||||
tf.config.experimental.set_memory_growth(device, True)
|
||||
except:
|
||||
self.logger.warning("no GPU device available")
|
||||
|
||||
self.logger.info("Loading models...")
|
||||
self.setup_models()
|
||||
self.logger.info(f"Model initialization complete ({time.time() - t_start:.1f}s)")
|
||||
|
||||
def setup_models(self):
|
||||
|
||||
loadable = [
|
||||
"col_classifier",
|
||||
"binarization",
|
||||
"page",
|
||||
"extract_images",
|
||||
]
|
||||
self.model_zoo.load_models(*loadable)
|
||||
|
||||
def get_regions_light_v_extract_only_images(self,img, num_col_classifier):
|
||||
self.logger.debug("enter get_regions_extract_images_only")
|
||||
erosion_hurts = False
|
||||
img_org = np.copy(img)
|
||||
img_height_h = img_org.shape[0]
|
||||
img_width_h = img_org.shape[1]
|
||||
|
||||
if num_col_classifier == 1:
|
||||
img_w_new = 700
|
||||
elif num_col_classifier == 2:
|
||||
img_w_new = 900
|
||||
elif num_col_classifier == 3:
|
||||
img_w_new = 1500
|
||||
elif num_col_classifier == 4:
|
||||
img_w_new = 1800
|
||||
elif num_col_classifier == 5:
|
||||
img_w_new = 2200
|
||||
elif num_col_classifier == 6:
|
||||
img_w_new = 2500
|
||||
else:
|
||||
raise ValueError("num_col_classifier must be in range 1..6")
|
||||
img_h_new = int(img.shape[0] / float(img.shape[1]) * img_w_new)
|
||||
img_resized = resize_image(img,img_h_new, img_w_new )
|
||||
|
||||
prediction_regions_org, _ = self.do_prediction_new_concept(True, img_resized, self.model_zoo.get("extract_images"))
|
||||
|
||||
prediction_regions_org = resize_image(prediction_regions_org,img_height_h, img_width_h )
|
||||
image_page, page_coord, cont_page = self.extract_page()
|
||||
|
||||
prediction_regions_org = prediction_regions_org[page_coord[0] : page_coord[1], page_coord[2] : page_coord[3]]
|
||||
prediction_regions_org=prediction_regions_org[:,:,0]
|
||||
|
||||
mask_lines_only = (prediction_regions_org[:,:] ==3)*1
|
||||
mask_texts_only = (prediction_regions_org[:,:] ==1)*1
|
||||
mask_images_only=(prediction_regions_org[:,:] ==2)*1
|
||||
|
||||
polygons_seplines, hir_seplines = return_contours_of_image(mask_lines_only)
|
||||
polygons_seplines = filter_contours_area_of_image(
|
||||
mask_lines_only, polygons_seplines, hir_seplines, max_area=1, min_area=0.00001, dilate=1)
|
||||
|
||||
polygons_of_only_texts = return_contours_of_interested_region(mask_texts_only,1,0.00001)
|
||||
polygons_of_only_lines = return_contours_of_interested_region(mask_lines_only,1,0.00001)
|
||||
|
||||
text_regions_p_true = np.zeros(prediction_regions_org.shape)
|
||||
text_regions_p_true = cv2.fillPoly(text_regions_p_true, pts = polygons_of_only_lines, color=(3,3,3))
|
||||
|
||||
text_regions_p_true[:,:][mask_images_only[:,:] == 1] = 2
|
||||
text_regions_p_true = cv2.fillPoly(text_regions_p_true, pts=polygons_of_only_texts, color=(1,1,1))
|
||||
|
||||
text_regions_p_true[text_regions_p_true.shape[0]-15:text_regions_p_true.shape[0], :] = 0
|
||||
text_regions_p_true[:, text_regions_p_true.shape[1]-15:text_regions_p_true.shape[1]] = 0
|
||||
|
||||
##polygons_of_images = return_contours_of_interested_region(text_regions_p_true, 2, 0.0001)
|
||||
polygons_of_images = return_contours_of_interested_region(text_regions_p_true, 2, 0.001)
|
||||
|
||||
polygons_of_images_fin = []
|
||||
for ploy_img_ind in polygons_of_images:
|
||||
box = _, _, w, h = cv2.boundingRect(ploy_img_ind)
|
||||
if h < 150 or w < 150:
|
||||
pass
|
||||
else:
|
||||
page_coord_img = box2rect(box) # type: ignore
|
||||
polygons_of_images_fin.append(np.array([[page_coord_img[2], page_coord_img[0]],
|
||||
[page_coord_img[3], page_coord_img[0]],
|
||||
[page_coord_img[3], page_coord_img[1]],
|
||||
[page_coord_img[2], page_coord_img[1]]]))
|
||||
|
||||
self.logger.debug("exit get_regions_extract_images_only")
|
||||
return (text_regions_p_true,
|
||||
erosion_hurts,
|
||||
polygons_seplines,
|
||||
polygons_of_images_fin,
|
||||
image_page,
|
||||
page_coord,
|
||||
cont_page)
|
||||
|
||||
def run(self,
|
||||
overwrite: bool = False,
|
||||
image_filename: Optional[str] = None,
|
||||
dir_in: Optional[str] = None,
|
||||
dir_out: Optional[str] = None,
|
||||
dir_of_cropped_images: Optional[str] = None,
|
||||
dir_of_layout: Optional[str] = None,
|
||||
dir_of_deskewed: Optional[str] = None,
|
||||
dir_of_all: Optional[str] = None,
|
||||
dir_save_page: Optional[str] = None,
|
||||
):
|
||||
"""
|
||||
Get image and scales, then extract the page of scanned image
|
||||
"""
|
||||
self.logger.debug("enter run")
|
||||
t0_tot = time.time()
|
||||
# Log enabled features directly
|
||||
enabled_modes = []
|
||||
if self.full_layout:
|
||||
enabled_modes.append("Full layout analysis")
|
||||
if self.tables:
|
||||
enabled_modes.append("Table detection")
|
||||
if enabled_modes:
|
||||
self.logger.info("Enabled modes: " + ", ".join(enabled_modes))
|
||||
if self.enable_plotting:
|
||||
self.logger.info("Saving debug plots")
|
||||
if dir_of_cropped_images:
|
||||
self.logger.info(f"Saving cropped images to: {dir_of_cropped_images}")
|
||||
if dir_of_layout:
|
||||
self.logger.info(f"Saving layout plots to: {dir_of_layout}")
|
||||
if dir_of_deskewed:
|
||||
self.logger.info(f"Saving deskewed images to: {dir_of_deskewed}")
|
||||
|
||||
if dir_in:
|
||||
ls_imgs = [os.path.join(dir_in, image_filename)
|
||||
for image_filename in filter(is_image_filename,
|
||||
os.listdir(dir_in))]
|
||||
elif image_filename:
|
||||
ls_imgs = [image_filename]
|
||||
else:
|
||||
raise ValueError("run requires either a single image filename or a directory")
|
||||
|
||||
for img_filename in ls_imgs:
|
||||
self.logger.info(img_filename)
|
||||
t0 = time.time()
|
||||
|
||||
self.reset_file_name_dir(img_filename, dir_out)
|
||||
if self.enable_plotting:
|
||||
self.plotter = EynollahPlotter(dir_out=dir_out,
|
||||
dir_of_all=dir_of_all,
|
||||
dir_save_page=dir_save_page,
|
||||
dir_of_deskewed=dir_of_deskewed,
|
||||
dir_of_cropped_images=dir_of_cropped_images,
|
||||
dir_of_layout=dir_of_layout,
|
||||
image_filename_stem=Path(img_filename).stem)
|
||||
#print("text region early -11 in %.1fs", time.time() - t0)
|
||||
if os.path.exists(self.writer.output_filename):
|
||||
if overwrite:
|
||||
self.logger.warning("will overwrite existing output file '%s'", self.writer.output_filename)
|
||||
else:
|
||||
self.logger.warning("will skip input for existing output file '%s'", self.writer.output_filename)
|
||||
continue
|
||||
|
||||
pcgts = self.run_single()
|
||||
self.logger.info("Job done in %.1fs", time.time() - t0)
|
||||
self.writer.write_pagexml(pcgts)
|
||||
|
||||
if dir_in:
|
||||
self.logger.info("All jobs done in %.1fs", time.time() - t0_tot)
|
||||
|
||||
def run_single(self):
|
||||
t0 = time.time()
|
||||
|
||||
self.logger.info(f"Processing file: {self.writer.image_filename}")
|
||||
self.logger.info("Step 1/5: Image Enhancement")
|
||||
|
||||
img_res, is_image_enhanced, num_col_classifier, _ = \
|
||||
self.run_enhancement()
|
||||
|
||||
self.logger.info(f"Image: {self.image.shape[1]}x{self.image.shape[0]}, "
|
||||
f"{self.dpi} DPI, {num_col_classifier} columns")
|
||||
if is_image_enhanced:
|
||||
self.logger.info("Enhancement applied")
|
||||
|
||||
self.logger.info(f"Enhancement complete ({time.time() - t0:.1f}s)")
|
||||
|
||||
|
||||
# Image Extraction Mode
|
||||
self.logger.info("Step 2/5: Image Extraction Mode")
|
||||
|
||||
_, _, _, polygons_of_images, \
|
||||
image_page, page_coord, cont_page = \
|
||||
self.get_regions_light_v_extract_only_images(img_res, num_col_classifier)
|
||||
|
||||
pcgts = self.writer.build_pagexml_no_full_layout(
|
||||
found_polygons_text_region=[],
|
||||
page_coord=page_coord,
|
||||
order_of_texts=[],
|
||||
all_found_textline_polygons=[],
|
||||
all_box_coord=[],
|
||||
found_polygons_text_region_img=polygons_of_images,
|
||||
found_polygons_marginals_left=[],
|
||||
found_polygons_marginals_right=[],
|
||||
all_found_textline_polygons_marginals_left=[],
|
||||
all_found_textline_polygons_marginals_right=[],
|
||||
all_box_coord_marginals_left=[],
|
||||
all_box_coord_marginals_right=[],
|
||||
slopes=[],
|
||||
slopes_marginals_left=[],
|
||||
slopes_marginals_right=[],
|
||||
cont_page=cont_page,
|
||||
polygons_seplines=[],
|
||||
found_polygons_tables=[],
|
||||
)
|
||||
if self.plotter:
|
||||
self.plotter.write_images_into_directory(polygons_of_images, image_page)
|
||||
|
||||
self.logger.info("Image extraction complete")
|
||||
return pcgts
|
||||
File diff suppressed because it is too large
Load diff
|
|
@ -1,10 +0,0 @@
|
|||
"""
|
||||
Load libraries with possible race conditions once. This must be imported as the first module of eynollah.
|
||||
"""
|
||||
from ocrd_utils import tf_disable_interactive_logs
|
||||
from torch import *
|
||||
tf_disable_interactive_logs()
|
||||
import tensorflow.keras
|
||||
from shapely import *
|
||||
imported_libs = True
|
||||
__all__ = ['imported_libs']
|
||||
|
|
@ -1,837 +0,0 @@
|
|||
# FIXME: fix all of those...
|
||||
# pyright: reportOptionalSubscript=false
|
||||
|
||||
from logging import Logger, getLogger
|
||||
from typing import List, Optional
|
||||
from pathlib import Path
|
||||
import os
|
||||
import gc
|
||||
import math
|
||||
from dataclasses import dataclass
|
||||
|
||||
import cv2
|
||||
from cv2.typing import MatLike
|
||||
from xml.etree import ElementTree as ET
|
||||
from PIL import Image, ImageDraw
|
||||
import numpy as np
|
||||
from eynollah.model_zoo import EynollahModelZoo
|
||||
from eynollah.utils.font import get_font
|
||||
from eynollah.utils.xml import etree_namespace_for_element_tag
|
||||
try:
|
||||
import torch
|
||||
except ImportError:
|
||||
torch = None
|
||||
|
||||
|
||||
from .utils import is_image_filename
|
||||
from .utils.resize import resize_image
|
||||
from .utils.utils_ocr import (
|
||||
break_curved_line_into_small_pieces_and_then_merge,
|
||||
decode_batch_predictions,
|
||||
fit_text_single_line,
|
||||
get_contours_and_bounding_boxes,
|
||||
get_orientation_moments,
|
||||
preprocess_and_resize_image_for_ocrcnn_model,
|
||||
return_textlines_split_if_needed,
|
||||
rotate_image_with_padding,
|
||||
)
|
||||
|
||||
# TODO: refine typing
|
||||
@dataclass
|
||||
class EynollahOcrResult:
|
||||
extracted_texts_merged: List
|
||||
extracted_conf_value_merged: Optional[List]
|
||||
cropped_lines_region_indexer: List
|
||||
total_bb_coordinates:List
|
||||
|
||||
class Eynollah_ocr:
|
||||
def __init__(
|
||||
self,
|
||||
*,
|
||||
model_zoo: EynollahModelZoo,
|
||||
tr_ocr=False,
|
||||
batch_size: Optional[int]=None,
|
||||
do_not_mask_with_textline_contour: bool=False,
|
||||
min_conf_value_of_textline_text : Optional[float]=None,
|
||||
logger: Optional[Logger]=None,
|
||||
):
|
||||
self.tr_ocr = tr_ocr
|
||||
# masking for OCR and GT generation, relevant for skewed lines and bounding boxes
|
||||
self.do_not_mask_with_textline_contour = do_not_mask_with_textline_contour
|
||||
self.logger = logger if logger else getLogger('eynollah.ocr')
|
||||
self.model_zoo = model_zoo
|
||||
|
||||
self.min_conf_value_of_textline_text = min_conf_value_of_textline_text if min_conf_value_of_textline_text else 0.3
|
||||
self.b_s = 2 if batch_size is None and tr_ocr else 8 if batch_size is None else batch_size
|
||||
|
||||
if tr_ocr:
|
||||
self.model_zoo.load_model('trocr_processor')
|
||||
self.model_zoo.load_model('ocr', 'tr')
|
||||
self.model_zoo.get('ocr').to(self.device)
|
||||
else:
|
||||
self.model_zoo.load_model('ocr', '')
|
||||
self.model_zoo.load_model('num_to_char')
|
||||
self.model_zoo.load_model('characters')
|
||||
self.end_character = len(self.model_zoo.get('characters', list)) + 2
|
||||
|
||||
@property
|
||||
def device(self):
|
||||
assert torch
|
||||
if torch.cuda.is_available():
|
||||
self.logger.info("Using GPU acceleration")
|
||||
return torch.device("cuda:0")
|
||||
else:
|
||||
self.logger.info("Using CPU processing")
|
||||
return torch.device("cpu")
|
||||
|
||||
def run_trocr(
|
||||
self,
|
||||
*,
|
||||
img: MatLike,
|
||||
page_tree: ET.ElementTree,
|
||||
page_ns,
|
||||
tr_ocr_input_height_and_width,
|
||||
) -> EynollahOcrResult:
|
||||
|
||||
total_bb_coordinates = []
|
||||
|
||||
|
||||
cropped_lines = []
|
||||
cropped_lines_region_indexer = []
|
||||
cropped_lines_meging_indexing = []
|
||||
|
||||
extracted_texts = []
|
||||
|
||||
indexer_text_region = 0
|
||||
indexer_b_s = 0
|
||||
|
||||
for nn in page_tree.getroot().iter(f'{{{page_ns}}}TextRegion'):
|
||||
for child_textregion in nn:
|
||||
if child_textregion.tag.endswith("TextLine"):
|
||||
|
||||
for child_textlines in child_textregion:
|
||||
if child_textlines.tag.endswith("Coords"):
|
||||
cropped_lines_region_indexer.append(indexer_text_region)
|
||||
p_h=child_textlines.attrib['points'].split(' ')
|
||||
textline_coords = np.array( [ [int(x.split(',')[0]),
|
||||
int(x.split(',')[1]) ]
|
||||
for x in p_h] )
|
||||
x,y,w,h = cv2.boundingRect(textline_coords)
|
||||
|
||||
total_bb_coordinates.append([x,y,w,h])
|
||||
|
||||
h2w_ratio = h/float(w)
|
||||
|
||||
img_poly_on_img = np.copy(img)
|
||||
mask_poly = np.zeros(img.shape)
|
||||
mask_poly = cv2.fillPoly(mask_poly, pts=[textline_coords], color=(1, 1, 1))
|
||||
|
||||
mask_poly = mask_poly[y:y+h, x:x+w, :]
|
||||
img_crop = img_poly_on_img[y:y+h, x:x+w, :]
|
||||
img_crop[mask_poly==0] = 255
|
||||
|
||||
self.logger.debug("processing %d lines for '%s'",
|
||||
len(cropped_lines), nn.attrib['id'])
|
||||
if h2w_ratio > 0.1:
|
||||
cropped_lines.append(resize_image(img_crop,
|
||||
tr_ocr_input_height_and_width,
|
||||
tr_ocr_input_height_and_width) )
|
||||
cropped_lines_meging_indexing.append(0)
|
||||
indexer_b_s+=1
|
||||
if indexer_b_s==self.b_s:
|
||||
imgs = cropped_lines[:]
|
||||
cropped_lines = []
|
||||
indexer_b_s = 0
|
||||
|
||||
pixel_values_merged = self.model_zoo.get('trocr_processor')(imgs, return_tensors="pt").pixel_values
|
||||
generated_ids_merged = self.model_zoo.get('ocr').generate(
|
||||
pixel_values_merged.to(self.device))
|
||||
generated_text_merged = self.model_zoo.get('trocr_processor').batch_decode(
|
||||
generated_ids_merged, skip_special_tokens=True)
|
||||
|
||||
extracted_texts = extracted_texts + generated_text_merged
|
||||
|
||||
else:
|
||||
splited_images, _ = return_textlines_split_if_needed(img_crop, None)
|
||||
#print(splited_images)
|
||||
if splited_images:
|
||||
cropped_lines.append(resize_image(splited_images[0],
|
||||
tr_ocr_input_height_and_width,
|
||||
tr_ocr_input_height_and_width))
|
||||
cropped_lines_meging_indexing.append(1)
|
||||
indexer_b_s+=1
|
||||
|
||||
if indexer_b_s==self.b_s:
|
||||
imgs = cropped_lines[:]
|
||||
cropped_lines = []
|
||||
indexer_b_s = 0
|
||||
|
||||
pixel_values_merged = self.model_zoo.get('trocr_processor')(imgs, return_tensors="pt").pixel_values
|
||||
generated_ids_merged = self.model_zoo.get('ocr').generate(
|
||||
pixel_values_merged.to(self.device))
|
||||
generated_text_merged = self.model_zoo.get('trocr_processor').batch_decode(
|
||||
generated_ids_merged, skip_special_tokens=True)
|
||||
|
||||
extracted_texts = extracted_texts + generated_text_merged
|
||||
|
||||
|
||||
cropped_lines.append(resize_image(splited_images[1],
|
||||
tr_ocr_input_height_and_width,
|
||||
tr_ocr_input_height_and_width))
|
||||
cropped_lines_meging_indexing.append(-1)
|
||||
indexer_b_s+=1
|
||||
|
||||
if indexer_b_s==self.b_s:
|
||||
imgs = cropped_lines[:]
|
||||
cropped_lines = []
|
||||
indexer_b_s = 0
|
||||
|
||||
pixel_values_merged = self.model_zoo.get('trocr_processor')(imgs, return_tensors="pt").pixel_values
|
||||
generated_ids_merged = self.model_zoo.get('ocr').generate(
|
||||
pixel_values_merged.to(self.device))
|
||||
generated_text_merged = self.model_zoo.get('trocr_processor').batch_decode(
|
||||
generated_ids_merged, skip_special_tokens=True)
|
||||
|
||||
extracted_texts = extracted_texts + generated_text_merged
|
||||
|
||||
else:
|
||||
cropped_lines.append(img_crop)
|
||||
cropped_lines_meging_indexing.append(0)
|
||||
indexer_b_s+=1
|
||||
|
||||
if indexer_b_s==self.b_s:
|
||||
imgs = cropped_lines[:]
|
||||
cropped_lines = []
|
||||
indexer_b_s = 0
|
||||
|
||||
pixel_values_merged = self.model_zoo.get('trocr_processor')(imgs, return_tensors="pt").pixel_values
|
||||
generated_ids_merged = self.model_zoo.get('ocr').generate(
|
||||
pixel_values_merged.to(self.device))
|
||||
generated_text_merged = self.model_zoo.get('trocr_processor').batch_decode(
|
||||
generated_ids_merged, skip_special_tokens=True)
|
||||
|
||||
extracted_texts = extracted_texts + generated_text_merged
|
||||
|
||||
|
||||
|
||||
indexer_text_region = indexer_text_region +1
|
||||
|
||||
if indexer_b_s!=0:
|
||||
imgs = cropped_lines[:]
|
||||
cropped_lines = []
|
||||
indexer_b_s = 0
|
||||
|
||||
pixel_values_merged = self.model_zoo.get('trocr_processor')(imgs, return_tensors="pt").pixel_values
|
||||
generated_ids_merged = self.model_zoo.get('ocr').generate(pixel_values_merged.to(self.device))
|
||||
generated_text_merged = self.model_zoo.get('trocr_processor').batch_decode(generated_ids_merged, skip_special_tokens=True)
|
||||
|
||||
extracted_texts = extracted_texts + generated_text_merged
|
||||
|
||||
####extracted_texts = []
|
||||
####n_iterations = math.ceil(len(cropped_lines) / self.b_s)
|
||||
|
||||
####for i in range(n_iterations):
|
||||
####if i==(n_iterations-1):
|
||||
####n_start = i*self.b_s
|
||||
####imgs = cropped_lines[n_start:]
|
||||
####else:
|
||||
####n_start = i*self.b_s
|
||||
####n_end = (i+1)*self.b_s
|
||||
####imgs = cropped_lines[n_start:n_end]
|
||||
####pixel_values_merged = self.model_zoo.get('trocr_processor')(imgs, return_tensors="pt").pixel_values
|
||||
####generated_ids_merged = self.model_ocr.generate(
|
||||
#### pixel_values_merged.to(self.device))
|
||||
####generated_text_merged = self.model_zoo.get('trocr_processor').batch_decode(
|
||||
#### generated_ids_merged, skip_special_tokens=True)
|
||||
|
||||
####extracted_texts = extracted_texts + generated_text_merged
|
||||
|
||||
del cropped_lines
|
||||
gc.collect()
|
||||
|
||||
extracted_texts_merged = [extracted_texts[ind]
|
||||
if cropped_lines_meging_indexing[ind]==0
|
||||
else extracted_texts[ind]+" "+extracted_texts[ind+1]
|
||||
if cropped_lines_meging_indexing[ind]==1
|
||||
else None
|
||||
for ind in range(len(cropped_lines_meging_indexing))]
|
||||
|
||||
extracted_texts_merged = [ind for ind in extracted_texts_merged if ind is not None]
|
||||
#print(extracted_texts_merged, len(extracted_texts_merged))
|
||||
|
||||
return EynollahOcrResult(
|
||||
extracted_texts_merged=extracted_texts_merged,
|
||||
extracted_conf_value_merged=None,
|
||||
cropped_lines_region_indexer=cropped_lines_region_indexer,
|
||||
total_bb_coordinates=total_bb_coordinates,
|
||||
)
|
||||
|
||||
def run_cnn(
|
||||
self,
|
||||
*,
|
||||
img: MatLike,
|
||||
img_bin: Optional[MatLike],
|
||||
page_tree: ET.ElementTree,
|
||||
page_ns,
|
||||
image_width,
|
||||
image_height,
|
||||
) -> EynollahOcrResult:
|
||||
|
||||
total_bb_coordinates = []
|
||||
|
||||
cropped_lines = []
|
||||
img_crop_bin = None
|
||||
imgs_bin = None
|
||||
imgs_bin_ver_flipped = None
|
||||
cropped_lines_bin = []
|
||||
cropped_lines_ver_index = []
|
||||
cropped_lines_region_indexer = []
|
||||
cropped_lines_meging_indexing = []
|
||||
|
||||
indexer_text_region = 0
|
||||
for nn in page_tree.getroot().iter(f'{{{page_ns}}}TextRegion'):
|
||||
try:
|
||||
type_textregion = nn.attrib['type']
|
||||
except:
|
||||
type_textregion = 'paragraph'
|
||||
for child_textregion in nn:
|
||||
if child_textregion.tag.endswith("TextLine"):
|
||||
for child_textlines in child_textregion:
|
||||
if child_textlines.tag.endswith("Coords"):
|
||||
cropped_lines_region_indexer.append(indexer_text_region)
|
||||
p_h=child_textlines.attrib['points'].split(' ')
|
||||
textline_coords = np.array( [ [int(x.split(',')[0]),
|
||||
int(x.split(',')[1]) ]
|
||||
for x in p_h] )
|
||||
|
||||
x,y,w,h = cv2.boundingRect(textline_coords)
|
||||
|
||||
angle_radians = math.atan2(h, w)
|
||||
# Convert to degrees
|
||||
angle_degrees = math.degrees(angle_radians)
|
||||
if type_textregion=='drop-capital':
|
||||
angle_degrees = 0
|
||||
|
||||
total_bb_coordinates.append([x,y,w,h])
|
||||
|
||||
w_scaled = w * image_height/float(h)
|
||||
|
||||
img_poly_on_img = np.copy(img)
|
||||
if img_bin:
|
||||
img_poly_on_img_bin = np.copy(img_bin)
|
||||
img_crop_bin = img_poly_on_img_bin[y:y+h, x:x+w, :]
|
||||
|
||||
mask_poly = np.zeros(img.shape)
|
||||
mask_poly = cv2.fillPoly(mask_poly, pts=[textline_coords], color=(1, 1, 1))
|
||||
|
||||
|
||||
mask_poly = mask_poly[y:y+h, x:x+w, :]
|
||||
img_crop = img_poly_on_img[y:y+h, x:x+w, :]
|
||||
|
||||
# print(file_name, angle_degrees, w*h,
|
||||
# mask_poly[:,:,0].sum(),
|
||||
# mask_poly[:,:,0].sum() /float(w*h) ,
|
||||
# 'didi')
|
||||
|
||||
if angle_degrees > 3:
|
||||
better_des_slope = get_orientation_moments(textline_coords)
|
||||
|
||||
img_crop = rotate_image_with_padding(img_crop, better_des_slope)
|
||||
if img_bin:
|
||||
img_crop_bin = rotate_image_with_padding(img_crop_bin, better_des_slope)
|
||||
|
||||
mask_poly = rotate_image_with_padding(mask_poly, better_des_slope)
|
||||
mask_poly = mask_poly.astype('uint8')
|
||||
|
||||
#new bounding box
|
||||
x_n, y_n, w_n, h_n = get_contours_and_bounding_boxes(mask_poly[:,:,0])
|
||||
|
||||
mask_poly = mask_poly[y_n:y_n+h_n, x_n:x_n+w_n, :]
|
||||
img_crop = img_crop[y_n:y_n+h_n, x_n:x_n+w_n, :]
|
||||
|
||||
if not self.do_not_mask_with_textline_contour:
|
||||
img_crop[mask_poly==0] = 255
|
||||
if img_bin:
|
||||
img_crop_bin = img_crop_bin[y_n:y_n+h_n, x_n:x_n+w_n, :]
|
||||
if not self.do_not_mask_with_textline_contour:
|
||||
img_crop_bin[mask_poly==0] = 255
|
||||
|
||||
if mask_poly[:,:,0].sum() /float(w_n*h_n) < 0.50 and w_scaled > 90:
|
||||
if img_bin:
|
||||
img_crop, img_crop_bin = \
|
||||
break_curved_line_into_small_pieces_and_then_merge(
|
||||
img_crop, mask_poly, img_crop_bin)
|
||||
else:
|
||||
img_crop, _ = \
|
||||
break_curved_line_into_small_pieces_and_then_merge(
|
||||
img_crop, mask_poly)
|
||||
|
||||
else:
|
||||
better_des_slope = 0
|
||||
if not self.do_not_mask_with_textline_contour:
|
||||
img_crop[mask_poly==0] = 255
|
||||
if img_bin:
|
||||
if not self.do_not_mask_with_textline_contour:
|
||||
img_crop_bin[mask_poly==0] = 255
|
||||
if type_textregion=='drop-capital':
|
||||
pass
|
||||
else:
|
||||
if mask_poly[:,:,0].sum() /float(w*h) < 0.50 and w_scaled > 90:
|
||||
if img_bin:
|
||||
img_crop, img_crop_bin = \
|
||||
break_curved_line_into_small_pieces_and_then_merge(
|
||||
img_crop, mask_poly, img_crop_bin)
|
||||
else:
|
||||
img_crop, _ = \
|
||||
break_curved_line_into_small_pieces_and_then_merge(
|
||||
img_crop, mask_poly)
|
||||
|
||||
if w_scaled < 750:#1.5*image_width:
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(
|
||||
img_crop, image_height, image_width)
|
||||
cropped_lines.append(img_fin)
|
||||
if abs(better_des_slope) > 45:
|
||||
cropped_lines_ver_index.append(1)
|
||||
else:
|
||||
cropped_lines_ver_index.append(0)
|
||||
|
||||
cropped_lines_meging_indexing.append(0)
|
||||
if img_bin:
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(
|
||||
img_crop_bin, image_height, image_width)
|
||||
cropped_lines_bin.append(img_fin)
|
||||
else:
|
||||
splited_images, splited_images_bin = return_textlines_split_if_needed(
|
||||
img_crop, img_crop_bin if img_bin else None)
|
||||
if splited_images:
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(
|
||||
splited_images[0], image_height, image_width)
|
||||
cropped_lines.append(img_fin)
|
||||
cropped_lines_meging_indexing.append(1)
|
||||
|
||||
if abs(better_des_slope) > 45:
|
||||
cropped_lines_ver_index.append(1)
|
||||
else:
|
||||
cropped_lines_ver_index.append(0)
|
||||
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(
|
||||
splited_images[1], image_height, image_width)
|
||||
|
||||
cropped_lines.append(img_fin)
|
||||
cropped_lines_meging_indexing.append(-1)
|
||||
|
||||
if abs(better_des_slope) > 45:
|
||||
cropped_lines_ver_index.append(1)
|
||||
else:
|
||||
cropped_lines_ver_index.append(0)
|
||||
|
||||
if img_bin:
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(
|
||||
splited_images_bin[0], image_height, image_width)
|
||||
cropped_lines_bin.append(img_fin)
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(
|
||||
splited_images_bin[1], image_height, image_width)
|
||||
cropped_lines_bin.append(img_fin)
|
||||
|
||||
else:
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(
|
||||
img_crop, image_height, image_width)
|
||||
cropped_lines.append(img_fin)
|
||||
cropped_lines_meging_indexing.append(0)
|
||||
|
||||
if abs(better_des_slope) > 45:
|
||||
cropped_lines_ver_index.append(1)
|
||||
else:
|
||||
cropped_lines_ver_index.append(0)
|
||||
|
||||
if img_bin:
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(
|
||||
img_crop_bin, image_height, image_width)
|
||||
cropped_lines_bin.append(img_fin)
|
||||
|
||||
|
||||
indexer_text_region = indexer_text_region +1
|
||||
|
||||
extracted_texts = []
|
||||
extracted_conf_value = []
|
||||
|
||||
n_iterations = math.ceil(len(cropped_lines) / self.b_s)
|
||||
|
||||
# FIXME: copy pasta
|
||||
for i in range(n_iterations):
|
||||
if i==(n_iterations-1):
|
||||
n_start = i*self.b_s
|
||||
imgs = cropped_lines[n_start:]
|
||||
imgs = np.array(imgs)
|
||||
imgs = imgs.reshape(imgs.shape[0], image_height, image_width, 3)
|
||||
|
||||
ver_imgs = np.array( cropped_lines_ver_index[n_start:] )
|
||||
indices_ver = np.where(ver_imgs == 1)[0]
|
||||
|
||||
#print(indices_ver, 'indices_ver')
|
||||
if len(indices_ver)>0:
|
||||
imgs_ver_flipped = imgs[indices_ver, : ,: ,:]
|
||||
imgs_ver_flipped = imgs_ver_flipped[:,::-1,::-1,:]
|
||||
#print(imgs_ver_flipped, 'imgs_ver_flipped')
|
||||
|
||||
else:
|
||||
imgs_ver_flipped = None
|
||||
|
||||
if img_bin:
|
||||
imgs_bin = cropped_lines_bin[n_start:]
|
||||
imgs_bin = np.array(imgs_bin)
|
||||
imgs_bin = imgs_bin.reshape(imgs_bin.shape[0], image_height, image_width, 3)
|
||||
|
||||
if len(indices_ver)>0:
|
||||
imgs_bin_ver_flipped = imgs_bin[indices_ver, : ,: ,:]
|
||||
imgs_bin_ver_flipped = imgs_bin_ver_flipped[:,::-1,::-1,:]
|
||||
#print(imgs_ver_flipped, 'imgs_ver_flipped')
|
||||
|
||||
else:
|
||||
imgs_bin_ver_flipped = None
|
||||
else:
|
||||
n_start = i*self.b_s
|
||||
n_end = (i+1)*self.b_s
|
||||
imgs = cropped_lines[n_start:n_end]
|
||||
imgs = np.array(imgs).reshape(self.b_s, image_height, image_width, 3)
|
||||
|
||||
ver_imgs = np.array( cropped_lines_ver_index[n_start:n_end] )
|
||||
indices_ver = np.where(ver_imgs == 1)[0]
|
||||
#print(indices_ver, 'indices_ver')
|
||||
|
||||
if len(indices_ver)>0:
|
||||
imgs_ver_flipped = imgs[indices_ver, : ,: ,:]
|
||||
imgs_ver_flipped = imgs_ver_flipped[:,::-1,::-1,:]
|
||||
#print(imgs_ver_flipped, 'imgs_ver_flipped')
|
||||
else:
|
||||
imgs_ver_flipped = None
|
||||
|
||||
|
||||
if img_bin:
|
||||
imgs_bin = cropped_lines_bin[n_start:n_end]
|
||||
imgs_bin = np.array(imgs_bin).reshape(self.b_s, image_height, image_width, 3)
|
||||
|
||||
|
||||
if len(indices_ver)>0:
|
||||
imgs_bin_ver_flipped = imgs_bin[indices_ver, : ,: ,:]
|
||||
imgs_bin_ver_flipped = imgs_bin_ver_flipped[:,::-1,::-1,:]
|
||||
#print(imgs_ver_flipped, 'imgs_ver_flipped')
|
||||
else:
|
||||
imgs_bin_ver_flipped = None
|
||||
|
||||
|
||||
self.logger.debug("processing next %d lines", len(imgs))
|
||||
preds = self.model_zoo.get('ocr').predict(imgs, verbose=0)
|
||||
|
||||
if len(indices_ver)>0:
|
||||
preds_flipped = self.model_zoo.get('ocr').predict(imgs_ver_flipped, verbose=0)
|
||||
preds_max_fliped = np.max(preds_flipped, axis=2 )
|
||||
preds_max_args_flipped = np.argmax(preds_flipped, axis=2 )
|
||||
pred_max_not_unk_mask_bool_flipped = preds_max_args_flipped[:,:]!=self.end_character
|
||||
masked_means_flipped = \
|
||||
np.sum(preds_max_fliped * pred_max_not_unk_mask_bool_flipped, axis=1) / \
|
||||
np.sum(pred_max_not_unk_mask_bool_flipped, axis=1)
|
||||
masked_means_flipped[np.isnan(masked_means_flipped)] = 0
|
||||
|
||||
preds_max = np.max(preds, axis=2 )
|
||||
preds_max_args = np.argmax(preds, axis=2 )
|
||||
pred_max_not_unk_mask_bool = preds_max_args[:,:]!=self.end_character
|
||||
|
||||
masked_means = \
|
||||
np.sum(preds_max * pred_max_not_unk_mask_bool, axis=1) / \
|
||||
np.sum(pred_max_not_unk_mask_bool, axis=1)
|
||||
masked_means[np.isnan(masked_means)] = 0
|
||||
|
||||
masked_means_ver = masked_means[indices_ver]
|
||||
#print(masked_means_ver, 'pred_max_not_unk')
|
||||
|
||||
indices_where_flipped_conf_value_is_higher = \
|
||||
np.where(masked_means_flipped > masked_means_ver)[0]
|
||||
|
||||
#print(indices_where_flipped_conf_value_is_higher, 'indices_where_flipped_conf_value_is_higher')
|
||||
if len(indices_where_flipped_conf_value_is_higher)>0:
|
||||
indices_to_be_replaced = indices_ver[indices_where_flipped_conf_value_is_higher]
|
||||
preds[indices_to_be_replaced,:,:] = \
|
||||
preds_flipped[indices_where_flipped_conf_value_is_higher, :, :]
|
||||
|
||||
if img_bin:
|
||||
preds_bin = self.model_zoo.get('ocr').predict(imgs_bin, verbose=0)
|
||||
|
||||
if len(indices_ver)>0:
|
||||
preds_flipped = self.model_zoo.get('ocr').predict(imgs_bin_ver_flipped, verbose=0)
|
||||
preds_max_fliped = np.max(preds_flipped, axis=2 )
|
||||
preds_max_args_flipped = np.argmax(preds_flipped, axis=2 )
|
||||
pred_max_not_unk_mask_bool_flipped = preds_max_args_flipped[:,:]!=self.end_character
|
||||
masked_means_flipped = \
|
||||
np.sum(preds_max_fliped * pred_max_not_unk_mask_bool_flipped, axis=1) / \
|
||||
np.sum(pred_max_not_unk_mask_bool_flipped, axis=1)
|
||||
masked_means_flipped[np.isnan(masked_means_flipped)] = 0
|
||||
|
||||
preds_max = np.max(preds, axis=2 )
|
||||
preds_max_args = np.argmax(preds, axis=2 )
|
||||
pred_max_not_unk_mask_bool = preds_max_args[:,:]!=self.end_character
|
||||
|
||||
masked_means = \
|
||||
np.sum(preds_max * pred_max_not_unk_mask_bool, axis=1) / \
|
||||
np.sum(pred_max_not_unk_mask_bool, axis=1)
|
||||
masked_means[np.isnan(masked_means)] = 0
|
||||
|
||||
masked_means_ver = masked_means[indices_ver]
|
||||
#print(masked_means_ver, 'pred_max_not_unk')
|
||||
|
||||
indices_where_flipped_conf_value_is_higher = \
|
||||
np.where(masked_means_flipped > masked_means_ver)[0]
|
||||
|
||||
#print(indices_where_flipped_conf_value_is_higher, 'indices_where_flipped_conf_value_is_higher')
|
||||
if len(indices_where_flipped_conf_value_is_higher)>0:
|
||||
indices_to_be_replaced = indices_ver[indices_where_flipped_conf_value_is_higher]
|
||||
preds_bin[indices_to_be_replaced,:,:] = \
|
||||
preds_flipped[indices_where_flipped_conf_value_is_higher, :, :]
|
||||
|
||||
preds = (preds + preds_bin) / 2.
|
||||
|
||||
pred_texts = decode_batch_predictions(preds, self.model_zoo.get('num_to_char'))
|
||||
|
||||
preds_max = np.max(preds, axis=2 )
|
||||
preds_max_args = np.argmax(preds, axis=2 )
|
||||
pred_max_not_unk_mask_bool = preds_max_args[:,:]!=self.end_character
|
||||
masked_means = \
|
||||
np.sum(preds_max * pred_max_not_unk_mask_bool, axis=1) / \
|
||||
np.sum(pred_max_not_unk_mask_bool, axis=1)
|
||||
|
||||
for ib in range(imgs.shape[0]):
|
||||
pred_texts_ib = pred_texts[ib].replace("[UNK]", "")
|
||||
if masked_means[ib] >= self.min_conf_value_of_textline_text:
|
||||
extracted_texts.append(pred_texts_ib)
|
||||
extracted_conf_value.append(masked_means[ib])
|
||||
else:
|
||||
extracted_texts.append("")
|
||||
extracted_conf_value.append(0)
|
||||
del cropped_lines
|
||||
del cropped_lines_bin
|
||||
gc.collect()
|
||||
|
||||
extracted_texts_merged = [extracted_texts[ind]
|
||||
if cropped_lines_meging_indexing[ind]==0
|
||||
else extracted_texts[ind]+" "+extracted_texts[ind+1]
|
||||
if cropped_lines_meging_indexing[ind]==1
|
||||
else None
|
||||
for ind in range(len(cropped_lines_meging_indexing))]
|
||||
|
||||
extracted_conf_value_merged = [extracted_conf_value[ind] # type: ignore
|
||||
if cropped_lines_meging_indexing[ind]==0
|
||||
else (extracted_conf_value[ind]+extracted_conf_value[ind+1])/2.
|
||||
if cropped_lines_meging_indexing[ind]==1
|
||||
else None
|
||||
for ind in range(len(cropped_lines_meging_indexing))]
|
||||
|
||||
extracted_conf_value_merged: List[float] = [extracted_conf_value_merged[ind_cfm]
|
||||
for ind_cfm in range(len(extracted_texts_merged))
|
||||
if extracted_texts_merged[ind_cfm] is not None]
|
||||
|
||||
extracted_texts_merged = [ind for ind in extracted_texts_merged if ind is not None]
|
||||
|
||||
return EynollahOcrResult(
|
||||
extracted_texts_merged=extracted_texts_merged,
|
||||
extracted_conf_value_merged=extracted_conf_value_merged,
|
||||
cropped_lines_region_indexer=cropped_lines_region_indexer,
|
||||
total_bb_coordinates=total_bb_coordinates,
|
||||
)
|
||||
|
||||
def write_ocr(
|
||||
self,
|
||||
*,
|
||||
result: EynollahOcrResult,
|
||||
page_tree: ET.ElementTree,
|
||||
out_file_ocr,
|
||||
page_ns,
|
||||
img,
|
||||
out_image_with_text,
|
||||
):
|
||||
cropped_lines_region_indexer = result.cropped_lines_region_indexer
|
||||
total_bb_coordinates = result.total_bb_coordinates
|
||||
extracted_texts_merged = result.extracted_texts_merged
|
||||
extracted_conf_value_merged = result.extracted_conf_value_merged
|
||||
|
||||
unique_cropped_lines_region_indexer = np.unique(cropped_lines_region_indexer)
|
||||
if out_image_with_text:
|
||||
image_text = Image.new("RGB", (img.shape[1], img.shape[0]), "white")
|
||||
draw = ImageDraw.Draw(image_text)
|
||||
font = get_font()
|
||||
|
||||
for indexer_text, bb_ind in enumerate(total_bb_coordinates):
|
||||
x_bb = bb_ind[0]
|
||||
y_bb = bb_ind[1]
|
||||
w_bb = bb_ind[2]
|
||||
h_bb = bb_ind[3]
|
||||
|
||||
font = fit_text_single_line(draw, extracted_texts_merged[indexer_text],
|
||||
font.path, w_bb, int(h_bb*0.4) )
|
||||
|
||||
##draw.rectangle([x_bb, y_bb, x_bb + w_bb, y_bb + h_bb], outline="red", width=2)
|
||||
|
||||
text_bbox = draw.textbbox((0, 0), extracted_texts_merged[indexer_text], font=font)
|
||||
text_width = text_bbox[2] - text_bbox[0]
|
||||
text_height = text_bbox[3] - text_bbox[1]
|
||||
|
||||
text_x = x_bb + (w_bb - text_width) // 2 # Center horizontally
|
||||
text_y = y_bb + (h_bb - text_height) // 2 # Center vertically
|
||||
|
||||
# Draw the text
|
||||
draw.text((text_x, text_y), extracted_texts_merged[indexer_text], fill="black", font=font)
|
||||
image_text.save(out_image_with_text)
|
||||
|
||||
text_by_textregion = []
|
||||
for ind in unique_cropped_lines_region_indexer:
|
||||
ind = np.array(cropped_lines_region_indexer)==ind
|
||||
extracted_texts_merged_un = np.array(extracted_texts_merged)[ind]
|
||||
if len(extracted_texts_merged_un)>1:
|
||||
text_by_textregion_ind = ""
|
||||
next_glue = ""
|
||||
for indt in range(len(extracted_texts_merged_un)):
|
||||
if (extracted_texts_merged_un[indt].endswith('⸗') or
|
||||
extracted_texts_merged_un[indt].endswith('-') or
|
||||
extracted_texts_merged_un[indt].endswith('¬')):
|
||||
text_by_textregion_ind += next_glue + extracted_texts_merged_un[indt][:-1]
|
||||
next_glue = ""
|
||||
else:
|
||||
text_by_textregion_ind += next_glue + extracted_texts_merged_un[indt]
|
||||
next_glue = " "
|
||||
text_by_textregion.append(text_by_textregion_ind)
|
||||
else:
|
||||
text_by_textregion.append(" ".join(extracted_texts_merged_un))
|
||||
|
||||
indexer = 0
|
||||
indexer_textregion = 0
|
||||
for nn in page_tree.getroot().iter(f'{{{page_ns}}}TextRegion'):
|
||||
|
||||
is_textregion_text = False
|
||||
for childtest in nn:
|
||||
if childtest.tag.endswith("TextEquiv"):
|
||||
is_textregion_text = True
|
||||
|
||||
if not is_textregion_text:
|
||||
text_subelement_textregion = ET.SubElement(nn, 'TextEquiv')
|
||||
unicode_textregion = ET.SubElement(text_subelement_textregion, 'Unicode')
|
||||
|
||||
|
||||
has_textline = False
|
||||
for child_textregion in nn:
|
||||
if child_textregion.tag.endswith("TextLine"):
|
||||
|
||||
is_textline_text = False
|
||||
for childtest2 in child_textregion:
|
||||
if childtest2.tag.endswith("TextEquiv"):
|
||||
is_textline_text = True
|
||||
|
||||
|
||||
if not is_textline_text:
|
||||
text_subelement = ET.SubElement(child_textregion, 'TextEquiv')
|
||||
if extracted_conf_value_merged:
|
||||
text_subelement.set('conf', f"{extracted_conf_value_merged[indexer]:.2f}")
|
||||
unicode_textline = ET.SubElement(text_subelement, 'Unicode')
|
||||
unicode_textline.text = extracted_texts_merged[indexer]
|
||||
else:
|
||||
for childtest3 in child_textregion:
|
||||
if childtest3.tag.endswith("TextEquiv"):
|
||||
for child_uc in childtest3:
|
||||
if child_uc.tag.endswith("Unicode"):
|
||||
if extracted_conf_value_merged:
|
||||
childtest3.set('conf', f"{extracted_conf_value_merged[indexer]:.2f}")
|
||||
child_uc.text = extracted_texts_merged[indexer]
|
||||
|
||||
indexer = indexer + 1
|
||||
has_textline = True
|
||||
if has_textline:
|
||||
if is_textregion_text:
|
||||
for child4 in nn:
|
||||
if child4.tag.endswith("TextEquiv"):
|
||||
for childtr_uc in child4:
|
||||
if childtr_uc.tag.endswith("Unicode"):
|
||||
childtr_uc.text = text_by_textregion[indexer_textregion]
|
||||
else:
|
||||
unicode_textregion.text = text_by_textregion[indexer_textregion]
|
||||
indexer_textregion = indexer_textregion + 1
|
||||
|
||||
ET.register_namespace("",page_ns)
|
||||
page_tree.write(out_file_ocr, xml_declaration=True, method='xml', encoding="utf-8", default_namespace=None)
|
||||
|
||||
def run(
|
||||
self,
|
||||
*,
|
||||
overwrite: bool = False,
|
||||
dir_in: Optional[str] = None,
|
||||
dir_in_bin: Optional[str] = None,
|
||||
image_filename: Optional[str] = None,
|
||||
dir_xmls: str,
|
||||
dir_out_image_text: Optional[str] = None,
|
||||
dir_out: str,
|
||||
):
|
||||
"""
|
||||
Run OCR.
|
||||
|
||||
Args:
|
||||
|
||||
dir_in_bin (str): Prediction with RGB and binarized images for selected pages, should not be the default
|
||||
"""
|
||||
if dir_in:
|
||||
ls_imgs = [os.path.join(dir_in, image_filename)
|
||||
for image_filename in filter(is_image_filename,
|
||||
os.listdir(dir_in))]
|
||||
else:
|
||||
assert image_filename
|
||||
ls_imgs = [image_filename]
|
||||
|
||||
for img_filename in ls_imgs:
|
||||
file_stem = Path(img_filename).stem
|
||||
page_file_in = os.path.join(dir_xmls, file_stem+'.xml')
|
||||
out_file_ocr = os.path.join(dir_out, file_stem+'.xml')
|
||||
|
||||
if os.path.exists(out_file_ocr):
|
||||
if overwrite:
|
||||
self.logger.warning("will overwrite existing output file '%s'", out_file_ocr)
|
||||
else:
|
||||
self.logger.warning("will skip input for existing output file '%s'", out_file_ocr)
|
||||
return
|
||||
|
||||
img = cv2.imread(img_filename)
|
||||
|
||||
page_tree = ET.parse(page_file_in, parser = ET.XMLParser(encoding="utf-8"))
|
||||
page_ns = etree_namespace_for_element_tag(page_tree.getroot().tag)
|
||||
|
||||
out_image_with_text = None
|
||||
if dir_out_image_text:
|
||||
out_image_with_text = os.path.join(dir_out_image_text, file_stem + '.png')
|
||||
|
||||
img_bin = None
|
||||
if dir_in_bin:
|
||||
img_bin = cv2.imread(os.path.join(dir_in_bin, file_stem+'.png'))
|
||||
|
||||
|
||||
if self.tr_ocr:
|
||||
result = self.run_trocr(
|
||||
img=img,
|
||||
page_tree=page_tree,
|
||||
page_ns=page_ns,
|
||||
|
||||
tr_ocr_input_height_and_width = 384
|
||||
)
|
||||
else:
|
||||
result = self.run_cnn(
|
||||
img=img,
|
||||
page_tree=page_tree,
|
||||
page_ns=page_ns,
|
||||
|
||||
img_bin=img_bin,
|
||||
image_width=512,
|
||||
image_height=32,
|
||||
)
|
||||
|
||||
self.write_ocr(
|
||||
result=result,
|
||||
img=img,
|
||||
page_tree=page_tree,
|
||||
page_ns=page_ns,
|
||||
out_file_ocr=out_file_ocr,
|
||||
out_image_with_text=out_image_with_text,
|
||||
)
|
||||
|
|
@ -1,712 +0,0 @@
|
|||
"""
|
||||
Image enhancer. The output can be written as same scale of input or in new predicted scale.
|
||||
"""
|
||||
|
||||
# FIXME: fix all of those...
|
||||
# pyright: reportUnboundVariable=false
|
||||
# pyright: reportCallIssue=false
|
||||
# pyright: reportArgumentType=false
|
||||
|
||||
import logging
|
||||
import os
|
||||
import time
|
||||
from typing import Optional
|
||||
from pathlib import Path
|
||||
import gc
|
||||
|
||||
import cv2
|
||||
from keras.models import Model
|
||||
import numpy as np
|
||||
import tensorflow as tf # type: ignore
|
||||
from skimage.morphology import skeletonize
|
||||
|
||||
from .model_zoo import EynollahModelZoo
|
||||
from .utils.resize import resize_image
|
||||
from .utils.pil_cv2 import pil2cv
|
||||
from .utils import (
|
||||
is_image_filename,
|
||||
crop_image_inside_box
|
||||
)
|
||||
|
||||
DPI_THRESHOLD = 298
|
||||
KERNEL = np.ones((5, 5), np.uint8)
|
||||
|
||||
|
||||
class Enhancer:
|
||||
def __init__(
|
||||
self,
|
||||
*,
|
||||
model_zoo: EynollahModelZoo,
|
||||
num_col_upper : Optional[int] = None,
|
||||
num_col_lower : Optional[int] = None,
|
||||
save_org_scale : bool = False,
|
||||
):
|
||||
self.input_binary = False
|
||||
self.save_org_scale = save_org_scale
|
||||
if num_col_upper:
|
||||
self.num_col_upper = int(num_col_upper)
|
||||
else:
|
||||
self.num_col_upper = num_col_upper
|
||||
if num_col_lower:
|
||||
self.num_col_lower = int(num_col_lower)
|
||||
else:
|
||||
self.num_col_lower = num_col_lower
|
||||
|
||||
self.logger = logging.getLogger('eynollah.enhance')
|
||||
self.model_zoo = model_zoo
|
||||
for v in ['binarization', 'enhancement', 'col_classifier', 'page']:
|
||||
self.model_zoo.load_model(v)
|
||||
|
||||
try:
|
||||
for device in tf.config.list_physical_devices('GPU'):
|
||||
tf.config.experimental.set_memory_growth(device, True)
|
||||
except:
|
||||
self.logger.warning("no GPU device available")
|
||||
|
||||
def cache_images(self, image_filename=None, image_pil=None, dpi=None):
|
||||
ret = {}
|
||||
if image_filename:
|
||||
ret['img'] = cv2.imread(image_filename)
|
||||
self.dpi = 100
|
||||
else:
|
||||
ret['img'] = pil2cv(image_pil)
|
||||
self.dpi = 100
|
||||
ret['img_grayscale'] = cv2.cvtColor(ret['img'], cv2.COLOR_BGR2GRAY)
|
||||
for prefix in ('', '_grayscale'):
|
||||
ret[f'img{prefix}_uint8'] = ret[f'img{prefix}'].astype(np.uint8)
|
||||
self._imgs = ret
|
||||
if dpi is not None:
|
||||
self.dpi = dpi
|
||||
|
||||
def reset_file_name_dir(self, image_filename, dir_out):
|
||||
self.cache_images(image_filename=image_filename)
|
||||
self.output_filename = os.path.join(dir_out, Path(image_filename).stem +'.png')
|
||||
|
||||
def imread(self, grayscale=False, uint8=True):
|
||||
key = 'img'
|
||||
if grayscale:
|
||||
key += '_grayscale'
|
||||
if uint8:
|
||||
key += '_uint8'
|
||||
return self._imgs[key].copy()
|
||||
|
||||
def predict_enhancement(self, img):
|
||||
self.logger.debug("enter predict_enhancement")
|
||||
|
||||
img_height_model = self.model_zoo.get('enhancement', Model).layers[-1].output_shape[1]
|
||||
img_width_model = self.model_zoo.get('enhancement', Model).layers[-1].output_shape[2]
|
||||
if img.shape[0] < img_height_model:
|
||||
img = cv2.resize(img, (img.shape[1], img_width_model), interpolation=cv2.INTER_NEAREST)
|
||||
if img.shape[1] < img_width_model:
|
||||
img = cv2.resize(img, (img_height_model, img.shape[0]), interpolation=cv2.INTER_NEAREST)
|
||||
margin = int(0.1 * img_width_model)
|
||||
width_mid = img_width_model - 2 * margin
|
||||
height_mid = img_height_model - 2 * margin
|
||||
img = img / 255.
|
||||
img_h = img.shape[0]
|
||||
img_w = img.shape[1]
|
||||
|
||||
prediction_true = np.zeros((img_h, img_w, 3))
|
||||
nxf = img_w / float(width_mid)
|
||||
nyf = img_h / float(height_mid)
|
||||
nxf = int(nxf) + 1 if nxf > int(nxf) else int(nxf)
|
||||
nyf = int(nyf) + 1 if nyf > int(nyf) else int(nyf)
|
||||
|
||||
for i in range(nxf):
|
||||
for j in range(nyf):
|
||||
if i == 0:
|
||||
index_x_d = i * width_mid
|
||||
index_x_u = index_x_d + img_width_model
|
||||
else:
|
||||
index_x_d = i * width_mid
|
||||
index_x_u = index_x_d + img_width_model
|
||||
if j == 0:
|
||||
index_y_d = j * height_mid
|
||||
index_y_u = index_y_d + img_height_model
|
||||
else:
|
||||
index_y_d = j * height_mid
|
||||
index_y_u = index_y_d + img_height_model
|
||||
|
||||
if index_x_u > img_w:
|
||||
index_x_u = img_w
|
||||
index_x_d = img_w - img_width_model
|
||||
if index_y_u > img_h:
|
||||
index_y_u = img_h
|
||||
index_y_d = img_h - img_height_model
|
||||
|
||||
img_patch = img[np.newaxis, index_y_d:index_y_u, index_x_d:index_x_u, :]
|
||||
label_p_pred = self.model_zoo.get('enhancement', Model).predict(img_patch, verbose='0')
|
||||
seg = label_p_pred[0, :, :, :] * 255
|
||||
|
||||
if i == 0 and j == 0:
|
||||
prediction_true[index_y_d + 0:index_y_u - margin,
|
||||
index_x_d + 0:index_x_u - margin] = \
|
||||
seg[0:-margin or None,
|
||||
0:-margin or None]
|
||||
elif i == nxf - 1 and j == nyf - 1:
|
||||
prediction_true[index_y_d + margin:index_y_u - 0,
|
||||
index_x_d + margin:index_x_u - 0] = \
|
||||
seg[margin:,
|
||||
margin:]
|
||||
elif i == 0 and j == nyf - 1:
|
||||
prediction_true[index_y_d + margin:index_y_u - 0,
|
||||
index_x_d + 0:index_x_u - margin] = \
|
||||
seg[margin:,
|
||||
0:-margin or None]
|
||||
elif i == nxf - 1 and j == 0:
|
||||
prediction_true[index_y_d + 0:index_y_u - margin,
|
||||
index_x_d + margin:index_x_u - 0] = \
|
||||
seg[0:-margin or None,
|
||||
margin:]
|
||||
elif i == 0 and j != 0 and j != nyf - 1:
|
||||
prediction_true[index_y_d + margin:index_y_u - margin,
|
||||
index_x_d + 0:index_x_u - margin] = \
|
||||
seg[margin:-margin or None,
|
||||
0:-margin or None]
|
||||
elif i == nxf - 1 and j != 0 and j != nyf - 1:
|
||||
prediction_true[index_y_d + margin:index_y_u - margin,
|
||||
index_x_d + margin:index_x_u - 0] = \
|
||||
seg[margin:-margin or None,
|
||||
margin:]
|
||||
elif i != 0 and i != nxf - 1 and j == 0:
|
||||
prediction_true[index_y_d + 0:index_y_u - margin,
|
||||
index_x_d + margin:index_x_u - margin] = \
|
||||
seg[0:-margin or None,
|
||||
margin:-margin or None]
|
||||
elif i != 0 and i != nxf - 1 and j == nyf - 1:
|
||||
prediction_true[index_y_d + margin:index_y_u - 0,
|
||||
index_x_d + margin:index_x_u - margin] = \
|
||||
seg[margin:,
|
||||
margin:-margin or None]
|
||||
else:
|
||||
prediction_true[index_y_d + margin:index_y_u - margin,
|
||||
index_x_d + margin:index_x_u - margin] = \
|
||||
seg[margin:-margin or None,
|
||||
margin:-margin or None]
|
||||
|
||||
prediction_true = prediction_true.astype(int)
|
||||
return prediction_true
|
||||
|
||||
def calculate_width_height_by_columns(self, img, num_col, width_early, label_p_pred):
|
||||
self.logger.debug("enter calculate_width_height_by_columns")
|
||||
if num_col == 1:
|
||||
img_w_new = 2000
|
||||
elif num_col == 2:
|
||||
img_w_new = 2400
|
||||
elif num_col == 3:
|
||||
img_w_new = 3000
|
||||
elif num_col == 4:
|
||||
img_w_new = 4000
|
||||
elif num_col == 5:
|
||||
img_w_new = 5000
|
||||
elif num_col == 6:
|
||||
img_w_new = 6500
|
||||
else:
|
||||
img_w_new = width_early
|
||||
img_h_new = img_w_new * img.shape[0] // img.shape[1]
|
||||
|
||||
if img_h_new >= 8000:
|
||||
img_new = np.copy(img)
|
||||
num_column_is_classified = False
|
||||
else:
|
||||
img_new = resize_image(img, img_h_new, img_w_new)
|
||||
num_column_is_classified = True
|
||||
|
||||
return img_new, num_column_is_classified
|
||||
|
||||
def early_page_for_num_of_column_classification(self,img_bin):
|
||||
self.logger.debug("enter early_page_for_num_of_column_classification")
|
||||
if self.input_binary:
|
||||
img = np.copy(img_bin).astype(np.uint8)
|
||||
else:
|
||||
img = self.imread()
|
||||
img = cv2.GaussianBlur(img, (5, 5), 0)
|
||||
img_page_prediction = self.do_prediction(False, img, self.model_zoo.get('page'))
|
||||
|
||||
imgray = cv2.cvtColor(img_page_prediction, cv2.COLOR_BGR2GRAY)
|
||||
_, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
thresh = cv2.dilate(thresh, KERNEL, iterations=3)
|
||||
contours, _ = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
if len(contours)>0:
|
||||
cnt_size = np.array([cv2.contourArea(contours[j])
|
||||
for j in range(len(contours))])
|
||||
cnt = contours[np.argmax(cnt_size)]
|
||||
box = cv2.boundingRect(cnt)
|
||||
else:
|
||||
box = [0, 0, img.shape[1], img.shape[0]]
|
||||
cropped_page, page_coord = crop_image_inside_box(box, img)
|
||||
|
||||
self.logger.debug("exit early_page_for_num_of_column_classification")
|
||||
return cropped_page, page_coord
|
||||
|
||||
def calculate_width_height_by_columns_1_2(self, img, num_col, width_early, label_p_pred):
|
||||
self.logger.debug("enter calculate_width_height_by_columns")
|
||||
if num_col == 1:
|
||||
img_w_new = 1000
|
||||
else:
|
||||
img_w_new = 1300
|
||||
img_h_new = img_w_new * img.shape[0] // img.shape[1]
|
||||
|
||||
if label_p_pred[0][int(num_col - 1)] < 0.9 and img_w_new < width_early:
|
||||
img_new = np.copy(img)
|
||||
num_column_is_classified = False
|
||||
#elif label_p_pred[0][int(num_col - 1)] < 0.8 and img_h_new >= 8000:
|
||||
elif img_h_new >= 8000:
|
||||
img_new = np.copy(img)
|
||||
num_column_is_classified = False
|
||||
else:
|
||||
img_new = resize_image(img, img_h_new, img_w_new)
|
||||
num_column_is_classified = True
|
||||
|
||||
return img_new, num_column_is_classified
|
||||
|
||||
def resize_and_enhance_image_with_column_classifier(self):
|
||||
self.logger.debug("enter resize_and_enhance_image_with_column_classifier")
|
||||
dpi = 0#self.dpi
|
||||
self.logger.info("Detected %s DPI", dpi)
|
||||
if self.input_binary:
|
||||
img = self.imread()
|
||||
prediction_bin = self.do_prediction(True, img, self.model_zoo.get('binarization'), n_batch_inference=5)
|
||||
prediction_bin = 255 * (prediction_bin[:,:,0]==0)
|
||||
prediction_bin = np.repeat(prediction_bin[:, :, np.newaxis], 3, axis=2).astype(np.uint8)
|
||||
img= np.copy(prediction_bin)
|
||||
img_bin = prediction_bin
|
||||
else:
|
||||
img = self.imread()
|
||||
self.h_org, self.w_org = img.shape[:2]
|
||||
img_bin = None
|
||||
|
||||
width_early = img.shape[1]
|
||||
t1 = time.time()
|
||||
_, page_coord = self.early_page_for_num_of_column_classification(img_bin)
|
||||
|
||||
self.image_page_org_size = img[page_coord[0] : page_coord[1], page_coord[2] : page_coord[3], :]
|
||||
self.page_coord = page_coord
|
||||
|
||||
if self.num_col_upper and not self.num_col_lower:
|
||||
num_col = self.num_col_upper
|
||||
label_p_pred = [np.ones(6)]
|
||||
elif self.num_col_lower and not self.num_col_upper:
|
||||
num_col = self.num_col_lower
|
||||
label_p_pred = [np.ones(6)]
|
||||
elif not self.num_col_upper and not self.num_col_lower:
|
||||
if self.input_binary:
|
||||
img_in = np.copy(img)
|
||||
img_in = img_in / 255.0
|
||||
img_in = cv2.resize(img_in, (448, 448), interpolation=cv2.INTER_NEAREST)
|
||||
img_in = img_in.reshape(1, 448, 448, 3)
|
||||
else:
|
||||
img_1ch = self.imread(grayscale=True)
|
||||
width_early = img_1ch.shape[1]
|
||||
img_1ch = img_1ch[page_coord[0] : page_coord[1], page_coord[2] : page_coord[3]]
|
||||
|
||||
img_1ch = img_1ch / 255.0
|
||||
img_1ch = cv2.resize(img_1ch, (448, 448), interpolation=cv2.INTER_NEAREST)
|
||||
img_in = np.zeros((1, img_1ch.shape[0], img_1ch.shape[1], 3))
|
||||
img_in[0, :, :, 0] = img_1ch[:, :]
|
||||
img_in[0, :, :, 1] = img_1ch[:, :]
|
||||
img_in[0, :, :, 2] = img_1ch[:, :]
|
||||
|
||||
label_p_pred = self.model_zoo.get('col_classifier').predict(img_in, verbose=0)
|
||||
num_col = np.argmax(label_p_pred[0]) + 1
|
||||
elif (self.num_col_upper and self.num_col_lower) and (self.num_col_upper!=self.num_col_lower):
|
||||
if self.input_binary:
|
||||
img_in = np.copy(img)
|
||||
img_in = img_in / 255.0
|
||||
img_in = cv2.resize(img_in, (448, 448), interpolation=cv2.INTER_NEAREST)
|
||||
img_in = img_in.reshape(1, 448, 448, 3)
|
||||
else:
|
||||
img_1ch = self.imread(grayscale=True)
|
||||
width_early = img_1ch.shape[1]
|
||||
img_1ch = img_1ch[page_coord[0] : page_coord[1], page_coord[2] : page_coord[3]]
|
||||
|
||||
img_1ch = img_1ch / 255.0
|
||||
img_1ch = cv2.resize(img_1ch, (448, 448), interpolation=cv2.INTER_NEAREST)
|
||||
img_in = np.zeros((1, img_1ch.shape[0], img_1ch.shape[1], 3))
|
||||
img_in[0, :, :, 0] = img_1ch[:, :]
|
||||
img_in[0, :, :, 1] = img_1ch[:, :]
|
||||
img_in[0, :, :, 2] = img_1ch[:, :]
|
||||
|
||||
label_p_pred = self.model_zoo.get('col_classifier').predict(img_in, verbose=0)
|
||||
num_col = np.argmax(label_p_pred[0]) + 1
|
||||
|
||||
if num_col > self.num_col_upper:
|
||||
num_col = self.num_col_upper
|
||||
label_p_pred = [np.ones(6)]
|
||||
if num_col < self.num_col_lower:
|
||||
num_col = self.num_col_lower
|
||||
label_p_pred = [np.ones(6)]
|
||||
else:
|
||||
num_col = self.num_col_upper
|
||||
label_p_pred = [np.ones(6)]
|
||||
|
||||
self.logger.info("Found %d columns (%s)", num_col, np.around(label_p_pred, decimals=5))
|
||||
|
||||
if dpi < DPI_THRESHOLD:
|
||||
if num_col in (1,2):
|
||||
img_new, num_column_is_classified = self.calculate_width_height_by_columns_1_2(
|
||||
img, num_col, width_early, label_p_pred)
|
||||
else:
|
||||
img_new, num_column_is_classified = self.calculate_width_height_by_columns(
|
||||
img, num_col, width_early, label_p_pred)
|
||||
image_res = np.copy(img_new)
|
||||
is_image_enhanced = True
|
||||
|
||||
else:
|
||||
num_column_is_classified = True
|
||||
image_res = np.copy(img)
|
||||
is_image_enhanced = False
|
||||
|
||||
self.logger.debug("exit resize_and_enhance_image_with_column_classifier")
|
||||
return is_image_enhanced, img, image_res, num_col, num_column_is_classified, img_bin
|
||||
def do_prediction(
|
||||
self, patches, img, model,
|
||||
n_batch_inference=1, marginal_of_patch_percent=0.1,
|
||||
thresholding_for_some_classes_in_light_version=False,
|
||||
thresholding_for_artificial_class_in_light_version=False, thresholding_for_fl_light_version=False, threshold_art_class_textline=0.1):
|
||||
|
||||
self.logger.debug("enter do_prediction")
|
||||
img_height_model = model.layers[-1].output_shape[1]
|
||||
img_width_model = model.layers[-1].output_shape[2]
|
||||
|
||||
if not patches:
|
||||
img_h_page = img.shape[0]
|
||||
img_w_page = img.shape[1]
|
||||
img = img / float(255.0)
|
||||
img = resize_image(img, img_height_model, img_width_model)
|
||||
|
||||
label_p_pred = model.predict(img[np.newaxis], verbose=0)
|
||||
seg = np.argmax(label_p_pred, axis=3)[0]
|
||||
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
seg_art = label_p_pred[0,:,:,2]
|
||||
|
||||
seg_art[seg_art<threshold_art_class_textline] = 0
|
||||
seg_art[seg_art>0] =1
|
||||
|
||||
skeleton_art = skeletonize(seg_art)
|
||||
skeleton_art = skeleton_art*1
|
||||
|
||||
seg[skeleton_art==1]=2
|
||||
|
||||
if thresholding_for_fl_light_version:
|
||||
seg_header = label_p_pred[0,:,:,2]
|
||||
|
||||
seg_header[seg_header<0.2] = 0
|
||||
seg_header[seg_header>0] =1
|
||||
|
||||
seg[seg_header==1]=2
|
||||
|
||||
seg_color = np.repeat(seg[:, :, np.newaxis], 3, axis=2)
|
||||
prediction_true = resize_image(seg_color, img_h_page, img_w_page).astype(np.uint8)
|
||||
return prediction_true
|
||||
|
||||
if img.shape[0] < img_height_model:
|
||||
img = resize_image(img, img_height_model, img.shape[1])
|
||||
if img.shape[1] < img_width_model:
|
||||
img = resize_image(img, img.shape[0], img_width_model)
|
||||
|
||||
self.logger.debug("Patch size: %sx%s", img_height_model, img_width_model)
|
||||
margin = int(marginal_of_patch_percent * img_height_model)
|
||||
width_mid = img_width_model - 2 * margin
|
||||
height_mid = img_height_model - 2 * margin
|
||||
img = img / 255.
|
||||
#img = img.astype(np.float16)
|
||||
img_h = img.shape[0]
|
||||
img_w = img.shape[1]
|
||||
prediction_true = np.zeros((img_h, img_w, 3))
|
||||
mask_true = np.zeros((img_h, img_w))
|
||||
nxf = img_w / float(width_mid)
|
||||
nyf = img_h / float(height_mid)
|
||||
nxf = int(nxf) + 1 if nxf > int(nxf) else int(nxf)
|
||||
nyf = int(nyf) + 1 if nyf > int(nyf) else int(nyf)
|
||||
|
||||
list_i_s = []
|
||||
list_j_s = []
|
||||
list_x_u = []
|
||||
list_x_d = []
|
||||
list_y_u = []
|
||||
list_y_d = []
|
||||
|
||||
batch_indexer = 0
|
||||
img_patch = np.zeros((n_batch_inference, img_height_model, img_width_model, 3))
|
||||
for i in range(nxf):
|
||||
for j in range(nyf):
|
||||
if i == 0:
|
||||
index_x_d = i * width_mid
|
||||
index_x_u = index_x_d + img_width_model
|
||||
else:
|
||||
index_x_d = i * width_mid
|
||||
index_x_u = index_x_d + img_width_model
|
||||
if j == 0:
|
||||
index_y_d = j * height_mid
|
||||
index_y_u = index_y_d + img_height_model
|
||||
else:
|
||||
index_y_d = j * height_mid
|
||||
index_y_u = index_y_d + img_height_model
|
||||
if index_x_u > img_w:
|
||||
index_x_u = img_w
|
||||
index_x_d = img_w - img_width_model
|
||||
if index_y_u > img_h:
|
||||
index_y_u = img_h
|
||||
index_y_d = img_h - img_height_model
|
||||
|
||||
list_i_s.append(i)
|
||||
list_j_s.append(j)
|
||||
list_x_u.append(index_x_u)
|
||||
list_x_d.append(index_x_d)
|
||||
list_y_d.append(index_y_d)
|
||||
list_y_u.append(index_y_u)
|
||||
|
||||
img_patch[batch_indexer,:,:,:] = img[index_y_d:index_y_u, index_x_d:index_x_u, :]
|
||||
batch_indexer += 1
|
||||
|
||||
if (batch_indexer == n_batch_inference or
|
||||
# last batch
|
||||
i == nxf - 1 and j == nyf - 1):
|
||||
self.logger.debug("predicting patches on %s", str(img_patch.shape))
|
||||
label_p_pred = model.predict(img_patch, verbose=0)
|
||||
seg = np.argmax(label_p_pred, axis=3)
|
||||
|
||||
if thresholding_for_some_classes_in_light_version:
|
||||
seg_not_base = label_p_pred[:,:,:,4]
|
||||
seg_not_base[seg_not_base>0.03] =1
|
||||
seg_not_base[seg_not_base<1] =0
|
||||
|
||||
seg_line = label_p_pred[:,:,:,3]
|
||||
seg_line[seg_line>0.1] =1
|
||||
seg_line[seg_line<1] =0
|
||||
|
||||
seg_background = label_p_pred[:,:,:,0]
|
||||
seg_background[seg_background>0.25] =1
|
||||
seg_background[seg_background<1] =0
|
||||
|
||||
seg[seg_not_base==1]=4
|
||||
seg[seg_background==1]=0
|
||||
seg[(seg_line==1) & (seg==0)]=3
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
seg_art = label_p_pred[:,:,:,2]
|
||||
|
||||
seg_art[seg_art<threshold_art_class_textline] = 0
|
||||
seg_art[seg_art>0] =1
|
||||
|
||||
##seg[seg_art==1]=2
|
||||
|
||||
indexer_inside_batch = 0
|
||||
for i_batch, j_batch in zip(list_i_s, list_j_s):
|
||||
seg_in = seg[indexer_inside_batch]
|
||||
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
seg_in_art = seg_art[indexer_inside_batch]
|
||||
|
||||
index_y_u_in = list_y_u[indexer_inside_batch]
|
||||
index_y_d_in = list_y_d[indexer_inside_batch]
|
||||
|
||||
index_x_u_in = list_x_u[indexer_inside_batch]
|
||||
index_x_d_in = list_x_d[indexer_inside_batch]
|
||||
|
||||
if i_batch == 0 and j_batch == 0:
|
||||
prediction_true[index_y_d_in + 0:index_y_u_in - margin,
|
||||
index_x_d_in + 0:index_x_u_in - margin] = \
|
||||
seg_in[0:-margin or None,
|
||||
0:-margin or None,
|
||||
np.newaxis]
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
prediction_true[index_y_d_in + 0:index_y_u_in - margin,
|
||||
index_x_d_in + 0:index_x_u_in - margin, 1] = \
|
||||
seg_in_art[0:-margin or None,
|
||||
0:-margin or None]
|
||||
|
||||
elif i_batch == nxf - 1 and j_batch == nyf - 1:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - 0,
|
||||
index_x_d_in + margin:index_x_u_in - 0] = \
|
||||
seg_in[margin:,
|
||||
margin:,
|
||||
np.newaxis]
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - 0,
|
||||
index_x_d_in + margin:index_x_u_in - 0, 1] = \
|
||||
seg_in_art[margin:,
|
||||
margin:]
|
||||
|
||||
elif i_batch == 0 and j_batch == nyf - 1:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - 0,
|
||||
index_x_d_in + 0:index_x_u_in - margin] = \
|
||||
seg_in[margin:,
|
||||
0:-margin or None,
|
||||
np.newaxis]
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - 0,
|
||||
index_x_d_in + 0:index_x_u_in - margin, 1] = \
|
||||
seg_in_art[margin:,
|
||||
0:-margin or None]
|
||||
|
||||
elif i_batch == nxf - 1 and j_batch == 0:
|
||||
prediction_true[index_y_d_in + 0:index_y_u_in - margin,
|
||||
index_x_d_in + margin:index_x_u_in - 0] = \
|
||||
seg_in[0:-margin or None,
|
||||
margin:,
|
||||
np.newaxis]
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
prediction_true[index_y_d_in + 0:index_y_u_in - margin,
|
||||
index_x_d_in + margin:index_x_u_in - 0, 1] = \
|
||||
seg_in_art[0:-margin or None,
|
||||
margin:]
|
||||
|
||||
elif i_batch == 0 and j_batch != 0 and j_batch != nyf - 1:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - margin,
|
||||
index_x_d_in + 0:index_x_u_in - margin] = \
|
||||
seg_in[margin:-margin or None,
|
||||
0:-margin or None,
|
||||
np.newaxis]
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - margin,
|
||||
index_x_d_in + 0:index_x_u_in - margin, 1] = \
|
||||
seg_in_art[margin:-margin or None,
|
||||
0:-margin or None]
|
||||
|
||||
elif i_batch == nxf - 1 and j_batch != 0 and j_batch != nyf - 1:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - margin,
|
||||
index_x_d_in + margin:index_x_u_in - 0] = \
|
||||
seg_in[margin:-margin or None,
|
||||
margin:,
|
||||
np.newaxis]
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - margin,
|
||||
index_x_d_in + margin:index_x_u_in - 0, 1] = \
|
||||
seg_in_art[margin:-margin or None,
|
||||
margin:]
|
||||
|
||||
elif i_batch != 0 and i_batch != nxf - 1 and j_batch == 0:
|
||||
prediction_true[index_y_d_in + 0:index_y_u_in - margin,
|
||||
index_x_d_in + margin:index_x_u_in - margin] = \
|
||||
seg_in[0:-margin or None,
|
||||
margin:-margin or None,
|
||||
np.newaxis]
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
prediction_true[index_y_d_in + 0:index_y_u_in - margin,
|
||||
index_x_d_in + margin:index_x_u_in - margin, 1] = \
|
||||
seg_in_art[0:-margin or None,
|
||||
margin:-margin or None]
|
||||
|
||||
elif i_batch != 0 and i_batch != nxf - 1 and j_batch == nyf - 1:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - 0,
|
||||
index_x_d_in + margin:index_x_u_in - margin] = \
|
||||
seg_in[margin:,
|
||||
margin:-margin or None,
|
||||
np.newaxis]
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - 0,
|
||||
index_x_d_in + margin:index_x_u_in - margin, 1] = \
|
||||
seg_in_art[margin:,
|
||||
margin:-margin or None]
|
||||
|
||||
else:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - margin,
|
||||
index_x_d_in + margin:index_x_u_in - margin] = \
|
||||
seg_in[margin:-margin or None,
|
||||
margin:-margin or None,
|
||||
np.newaxis]
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
prediction_true[index_y_d_in + margin:index_y_u_in - margin,
|
||||
index_x_d_in + margin:index_x_u_in - margin, 1] = \
|
||||
seg_in_art[margin:-margin or None,
|
||||
margin:-margin or None]
|
||||
indexer_inside_batch += 1
|
||||
|
||||
|
||||
list_i_s = []
|
||||
list_j_s = []
|
||||
list_x_u = []
|
||||
list_x_d = []
|
||||
list_y_u = []
|
||||
list_y_d = []
|
||||
|
||||
batch_indexer = 0
|
||||
img_patch[:] = 0
|
||||
|
||||
prediction_true = prediction_true.astype(np.uint8)
|
||||
|
||||
if thresholding_for_artificial_class_in_light_version:
|
||||
kernel_min = np.ones((3, 3), np.uint8)
|
||||
prediction_true[:,:,0][prediction_true[:,:,0]==2] = 0
|
||||
|
||||
skeleton_art = skeletonize(prediction_true[:,:,1])
|
||||
skeleton_art = skeleton_art*1
|
||||
|
||||
skeleton_art = skeleton_art.astype('uint8')
|
||||
|
||||
skeleton_art = cv2.dilate(skeleton_art, kernel_min, iterations=1)
|
||||
|
||||
prediction_true[:,:,0][skeleton_art==1]=2
|
||||
#del model
|
||||
gc.collect()
|
||||
return prediction_true
|
||||
|
||||
def run_enhancement(self):
|
||||
t_in = time.time()
|
||||
self.logger.info("Resizing and enhancing image...")
|
||||
is_image_enhanced, img_org, img_res, num_col_classifier, num_column_is_classified, img_bin = \
|
||||
self.resize_and_enhance_image_with_column_classifier()
|
||||
|
||||
self.logger.info("Image was %senhanced.", '' if is_image_enhanced else 'not ')
|
||||
return img_res, is_image_enhanced, num_col_classifier, num_column_is_classified
|
||||
|
||||
|
||||
def run_single(self):
|
||||
t0 = time.time()
|
||||
img_res, is_image_enhanced, num_col_classifier, num_column_is_classified = self.run_enhancement()
|
||||
|
||||
return img_res, is_image_enhanced
|
||||
|
||||
|
||||
def run(self,
|
||||
overwrite: bool = False,
|
||||
image_filename: Optional[str] = None,
|
||||
dir_in: Optional[str] = None,
|
||||
dir_out: Optional[str] = None,
|
||||
):
|
||||
"""
|
||||
Get image and scales, then extract the page of scanned image
|
||||
"""
|
||||
self.logger.debug("enter run")
|
||||
t0_tot = time.time()
|
||||
|
||||
if dir_in:
|
||||
ls_imgs = [os.path.join(dir_in, image_filename)
|
||||
for image_filename in filter(is_image_filename,
|
||||
os.listdir(dir_in))]
|
||||
elif image_filename:
|
||||
ls_imgs = [image_filename]
|
||||
else:
|
||||
raise ValueError("run requires either a single image filename or a directory")
|
||||
|
||||
for img_filename in ls_imgs:
|
||||
self.logger.info(img_filename)
|
||||
t0 = time.time()
|
||||
|
||||
self.reset_file_name_dir(img_filename, dir_out)
|
||||
#print("text region early -11 in %.1fs", time.time() - t0)
|
||||
|
||||
if os.path.exists(self.output_filename):
|
||||
if overwrite:
|
||||
self.logger.warning("will overwrite existing output file '%s'", self.output_filename)
|
||||
else:
|
||||
self.logger.warning("will skip input for existing output file '%s'", self.output_filename)
|
||||
continue
|
||||
|
||||
did_resize = False
|
||||
image_enhanced, did_enhance = self.run_single()
|
||||
if self.save_org_scale:
|
||||
image_enhanced = resize_image(image_enhanced, self.h_org, self.w_org)
|
||||
did_resize = True
|
||||
|
||||
self.logger.info(
|
||||
"Image %s was %senhanced%s.",
|
||||
img_filename,
|
||||
'' if did_enhance else 'not ',
|
||||
'and resized' if did_resize else ''
|
||||
)
|
||||
|
||||
cv2.imwrite(self.output_filename, image_enhanced)
|
||||
|
||||
|
|
@ -1,804 +0,0 @@
|
|||
"""
|
||||
Machine learning based reading order detection
|
||||
"""
|
||||
|
||||
# pyright: reportCallIssue=false
|
||||
# pyright: reportUnboundVariable=false
|
||||
# pyright: reportArgumentType=false
|
||||
|
||||
import logging
|
||||
import os
|
||||
import time
|
||||
from typing import Optional
|
||||
from pathlib import Path
|
||||
import xml.etree.ElementTree as ET
|
||||
|
||||
import cv2
|
||||
from keras.models import Model
|
||||
import numpy as np
|
||||
import statistics
|
||||
import tensorflow as tf
|
||||
|
||||
from .model_zoo import EynollahModelZoo
|
||||
from .utils.resize import resize_image
|
||||
from .utils.contour import (
|
||||
find_new_features_of_contours,
|
||||
return_contours_of_image,
|
||||
return_parent_contours,
|
||||
)
|
||||
from .utils import is_xml_filename
|
||||
|
||||
DPI_THRESHOLD = 298
|
||||
KERNEL = np.ones((5, 5), np.uint8)
|
||||
|
||||
|
||||
class machine_based_reading_order_on_layout:
|
||||
def __init__(
|
||||
self,
|
||||
*,
|
||||
model_zoo: EynollahModelZoo,
|
||||
logger : Optional[logging.Logger] = None,
|
||||
):
|
||||
self.logger = logger or logging.getLogger('eynollah.mbreorder')
|
||||
self.model_zoo = model_zoo
|
||||
|
||||
try:
|
||||
for device in tf.config.list_physical_devices('GPU'):
|
||||
tf.config.experimental.set_memory_growth(device, True)
|
||||
except:
|
||||
self.logger.warning("no GPU device available")
|
||||
|
||||
self.model_zoo.load_model('reading_order')
|
||||
|
||||
def read_xml(self, xml_file):
|
||||
tree1 = ET.parse(xml_file, parser = ET.XMLParser(encoding='utf-8'))
|
||||
root1=tree1.getroot()
|
||||
alltags=[elem.tag for elem in root1.iter()]
|
||||
link=alltags[0].split('}')[0]+'}'
|
||||
|
||||
index_tot_regions = []
|
||||
tot_region_ref = []
|
||||
|
||||
y_len, x_len = 0, 0
|
||||
for jj in root1.iter(link+'Page'):
|
||||
y_len=int(jj.attrib['imageHeight'])
|
||||
x_len=int(jj.attrib['imageWidth'])
|
||||
|
||||
for jj in root1.iter(link+'RegionRefIndexed'):
|
||||
index_tot_regions.append(jj.attrib['index'])
|
||||
tot_region_ref.append(jj.attrib['regionRef'])
|
||||
|
||||
if (link+'PrintSpace' in alltags) or (link+'Border' in alltags):
|
||||
co_printspace = []
|
||||
if link+'PrintSpace' in alltags:
|
||||
region_tags_printspace = np.unique([x for x in alltags if x.endswith('PrintSpace')])
|
||||
else:
|
||||
region_tags_printspace = np.unique([x for x in alltags if x.endswith('Border')])
|
||||
|
||||
for tag in region_tags_printspace:
|
||||
if link+'PrintSpace' in alltags:
|
||||
tag_endings_printspace = ['}PrintSpace','}printspace']
|
||||
else:
|
||||
tag_endings_printspace = ['}Border','}border']
|
||||
|
||||
if tag.endswith(tag_endings_printspace[0]) or tag.endswith(tag_endings_printspace[1]):
|
||||
for nn in root1.iter(tag):
|
||||
c_t_in = []
|
||||
sumi = 0
|
||||
for vv in nn.iter():
|
||||
# check the format of coords
|
||||
if vv.tag == link + 'Coords':
|
||||
coords = bool(vv.attrib)
|
||||
if coords:
|
||||
p_h = vv.attrib['points'].split(' ')
|
||||
c_t_in.append(
|
||||
np.array([[int(x.split(',')[0]), int(x.split(',')[1])] for x in p_h]))
|
||||
break
|
||||
else:
|
||||
pass
|
||||
|
||||
if vv.tag == link + 'Point':
|
||||
c_t_in.append([int(float(vv.attrib['x'])), int(float(vv.attrib['y']))])
|
||||
sumi += 1
|
||||
elif vv.tag != link + 'Point' and sumi >= 1:
|
||||
break
|
||||
co_printspace.append(np.array(c_t_in))
|
||||
img_printspace = np.zeros( (y_len,x_len,3) )
|
||||
img_printspace=cv2.fillPoly(img_printspace, pts =co_printspace, color=(1,1,1))
|
||||
img_printspace = img_printspace.astype(np.uint8)
|
||||
|
||||
imgray = cv2.cvtColor(img_printspace, cv2.COLOR_BGR2GRAY)
|
||||
_, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
contours, _ = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
cnt_size = np.array([cv2.contourArea(contours[j]) for j in range(len(contours))])
|
||||
cnt = contours[np.argmax(cnt_size)]
|
||||
x, y, w, h = cv2.boundingRect(cnt)
|
||||
|
||||
bb_coord_printspace = [x, y, w, h]
|
||||
|
||||
else:
|
||||
bb_coord_printspace = None
|
||||
|
||||
|
||||
region_tags=np.unique([x for x in alltags if x.endswith('Region')])
|
||||
co_text_paragraph=[]
|
||||
co_text_drop=[]
|
||||
co_text_heading=[]
|
||||
co_text_header=[]
|
||||
co_text_marginalia=[]
|
||||
co_text_catch=[]
|
||||
co_text_page_number=[]
|
||||
co_text_signature_mark=[]
|
||||
co_sep=[]
|
||||
co_img=[]
|
||||
co_table=[]
|
||||
co_graphic=[]
|
||||
co_graphic_text_annotation=[]
|
||||
co_graphic_decoration=[]
|
||||
co_noise=[]
|
||||
|
||||
co_text_paragraph_text=[]
|
||||
co_text_drop_text=[]
|
||||
co_text_heading_text=[]
|
||||
co_text_header_text=[]
|
||||
co_text_marginalia_text=[]
|
||||
co_text_catch_text=[]
|
||||
co_text_page_number_text=[]
|
||||
co_text_signature_mark_text=[]
|
||||
co_sep_text=[]
|
||||
co_img_text=[]
|
||||
co_table_text=[]
|
||||
co_graphic_text=[]
|
||||
co_graphic_text_annotation_text=[]
|
||||
co_graphic_decoration_text=[]
|
||||
co_noise_text=[]
|
||||
|
||||
id_paragraph = []
|
||||
id_header = []
|
||||
id_heading = []
|
||||
id_marginalia = []
|
||||
|
||||
for tag in region_tags:
|
||||
if tag.endswith('}TextRegion') or tag.endswith('}Textregion'):
|
||||
for nn in root1.iter(tag):
|
||||
for child2 in nn:
|
||||
tag2 = child2.tag
|
||||
if tag2.endswith('}TextEquiv') or tag2.endswith('}TextEquiv'):
|
||||
for childtext2 in child2:
|
||||
if childtext2.tag.endswith('}Unicode') or childtext2.tag.endswith('}Unicode'):
|
||||
if "type" in nn.attrib and nn.attrib['type']=='drop-capital':
|
||||
co_text_drop_text.append(childtext2.text)
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='heading':
|
||||
co_text_heading_text.append(childtext2.text)
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='signature-mark':
|
||||
co_text_signature_mark_text.append(childtext2.text)
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='header':
|
||||
co_text_header_text.append(childtext2.text)
|
||||
###elif "type" in nn.attrib and nn.attrib['type']=='catch-word':
|
||||
###co_text_catch_text.append(childtext2.text)
|
||||
###elif "type" in nn.attrib and nn.attrib['type']=='page-number':
|
||||
###co_text_page_number_text.append(childtext2.text)
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='marginalia':
|
||||
co_text_marginalia_text.append(childtext2.text)
|
||||
else:
|
||||
co_text_paragraph_text.append(childtext2.text)
|
||||
c_t_in_drop=[]
|
||||
c_t_in_paragraph=[]
|
||||
c_t_in_heading=[]
|
||||
c_t_in_header=[]
|
||||
c_t_in_page_number=[]
|
||||
c_t_in_signature_mark=[]
|
||||
c_t_in_catch=[]
|
||||
c_t_in_marginalia=[]
|
||||
|
||||
|
||||
sumi=0
|
||||
for vv in nn.iter():
|
||||
# check the format of coords
|
||||
if vv.tag==link+'Coords':
|
||||
|
||||
coords=bool(vv.attrib)
|
||||
if coords:
|
||||
#print('birda1')
|
||||
p_h=vv.attrib['points'].split(' ')
|
||||
|
||||
|
||||
|
||||
if "type" in nn.attrib and nn.attrib['type']=='drop-capital':
|
||||
|
||||
c_t_in_drop.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='heading':
|
||||
##id_heading.append(nn.attrib['id'])
|
||||
c_t_in_heading.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
|
||||
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='signature-mark':
|
||||
|
||||
c_t_in_signature_mark.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
#print(c_t_in_paragraph)
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='header':
|
||||
#id_header.append(nn.attrib['id'])
|
||||
c_t_in_header.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
|
||||
|
||||
###elif "type" in nn.attrib and nn.attrib['type']=='catch-word':
|
||||
###c_t_in_catch.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
|
||||
|
||||
###elif "type" in nn.attrib and nn.attrib['type']=='page-number':
|
||||
|
||||
###c_t_in_page_number.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='marginalia':
|
||||
#id_marginalia.append(nn.attrib['id'])
|
||||
|
||||
c_t_in_marginalia.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
else:
|
||||
#id_paragraph.append(nn.attrib['id'])
|
||||
|
||||
c_t_in_paragraph.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
|
||||
break
|
||||
else:
|
||||
pass
|
||||
|
||||
|
||||
if vv.tag==link+'Point':
|
||||
if "type" in nn.attrib and nn.attrib['type']=='drop-capital':
|
||||
|
||||
c_t_in_drop.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='heading':
|
||||
#id_heading.append(nn.attrib['id'])
|
||||
c_t_in_heading.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
|
||||
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='signature-mark':
|
||||
|
||||
c_t_in_signature_mark.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='header':
|
||||
#id_header.append(nn.attrib['id'])
|
||||
c_t_in_header.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
|
||||
|
||||
###elif "type" in nn.attrib and nn.attrib['type']=='catch-word':
|
||||
###c_t_in_catch.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
###sumi+=1
|
||||
|
||||
###elif "type" in nn.attrib and nn.attrib['type']=='page-number':
|
||||
|
||||
###c_t_in_page_number.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
###sumi+=1
|
||||
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='marginalia':
|
||||
#id_marginalia.append(nn.attrib['id'])
|
||||
|
||||
c_t_in_marginalia.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
|
||||
else:
|
||||
#id_paragraph.append(nn.attrib['id'])
|
||||
c_t_in_paragraph.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
|
||||
elif vv.tag!=link+'Point' and sumi>=1:
|
||||
break
|
||||
|
||||
if len(c_t_in_drop)>0:
|
||||
co_text_drop.append(np.array(c_t_in_drop))
|
||||
if len(c_t_in_paragraph)>0:
|
||||
co_text_paragraph.append(np.array(c_t_in_paragraph))
|
||||
id_paragraph.append(nn.attrib['id'])
|
||||
if len(c_t_in_heading)>0:
|
||||
co_text_heading.append(np.array(c_t_in_heading))
|
||||
id_heading.append(nn.attrib['id'])
|
||||
|
||||
if len(c_t_in_header)>0:
|
||||
co_text_header.append(np.array(c_t_in_header))
|
||||
id_header.append(nn.attrib['id'])
|
||||
if len(c_t_in_page_number)>0:
|
||||
co_text_page_number.append(np.array(c_t_in_page_number))
|
||||
if len(c_t_in_catch)>0:
|
||||
co_text_catch.append(np.array(c_t_in_catch))
|
||||
|
||||
if len(c_t_in_signature_mark)>0:
|
||||
co_text_signature_mark.append(np.array(c_t_in_signature_mark))
|
||||
|
||||
if len(c_t_in_marginalia)>0:
|
||||
co_text_marginalia.append(np.array(c_t_in_marginalia))
|
||||
id_marginalia.append(nn.attrib['id'])
|
||||
|
||||
|
||||
elif tag.endswith('}GraphicRegion') or tag.endswith('}graphicregion'):
|
||||
for nn in root1.iter(tag):
|
||||
c_t_in=[]
|
||||
c_t_in_text_annotation=[]
|
||||
c_t_in_decoration=[]
|
||||
sumi=0
|
||||
for vv in nn.iter():
|
||||
# check the format of coords
|
||||
if vv.tag==link+'Coords':
|
||||
coords=bool(vv.attrib)
|
||||
if coords:
|
||||
p_h=vv.attrib['points'].split(' ')
|
||||
|
||||
if "type" in nn.attrib and nn.attrib['type']=='handwritten-annotation':
|
||||
c_t_in_text_annotation.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='decoration':
|
||||
c_t_in_decoration.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
|
||||
else:
|
||||
c_t_in.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
|
||||
|
||||
break
|
||||
else:
|
||||
pass
|
||||
|
||||
|
||||
if vv.tag==link+'Point':
|
||||
if "type" in nn.attrib and nn.attrib['type']=='handwritten-annotation':
|
||||
c_t_in_text_annotation.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
|
||||
elif "type" in nn.attrib and nn.attrib['type']=='decoration':
|
||||
c_t_in_decoration.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
|
||||
else:
|
||||
c_t_in.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
|
||||
if len(c_t_in_text_annotation)>0:
|
||||
co_graphic_text_annotation.append(np.array(c_t_in_text_annotation))
|
||||
if len(c_t_in_decoration)>0:
|
||||
co_graphic_decoration.append(np.array(c_t_in_decoration))
|
||||
if len(c_t_in)>0:
|
||||
co_graphic.append(np.array(c_t_in))
|
||||
|
||||
|
||||
|
||||
elif tag.endswith('}ImageRegion') or tag.endswith('}imageregion'):
|
||||
for nn in root1.iter(tag):
|
||||
c_t_in=[]
|
||||
sumi=0
|
||||
for vv in nn.iter():
|
||||
# check the format of coords
|
||||
if vv.tag==link+'Coords':
|
||||
coords=bool(vv.attrib)
|
||||
if coords:
|
||||
p_h=vv.attrib['points'].split(' ')
|
||||
c_t_in.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
break
|
||||
else:
|
||||
pass
|
||||
|
||||
|
||||
if vv.tag==link+'Point':
|
||||
c_t_in.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
elif vv.tag!=link+'Point' and sumi>=1:
|
||||
break
|
||||
co_img.append(np.array(c_t_in))
|
||||
co_img_text.append(' ')
|
||||
|
||||
|
||||
elif tag.endswith('}SeparatorRegion') or tag.endswith('}separatorregion'):
|
||||
for nn in root1.iter(tag):
|
||||
c_t_in=[]
|
||||
sumi=0
|
||||
for vv in nn.iter():
|
||||
# check the format of coords
|
||||
if vv.tag==link+'Coords':
|
||||
coords=bool(vv.attrib)
|
||||
if coords:
|
||||
p_h=vv.attrib['points'].split(' ')
|
||||
c_t_in.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
break
|
||||
else:
|
||||
pass
|
||||
|
||||
|
||||
if vv.tag==link+'Point':
|
||||
c_t_in.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
elif vv.tag!=link+'Point' and sumi>=1:
|
||||
break
|
||||
co_sep.append(np.array(c_t_in))
|
||||
|
||||
|
||||
|
||||
elif tag.endswith('}TableRegion') or tag.endswith('}tableregion'):
|
||||
for nn in root1.iter(tag):
|
||||
c_t_in=[]
|
||||
sumi=0
|
||||
for vv in nn.iter():
|
||||
# check the format of coords
|
||||
if vv.tag==link+'Coords':
|
||||
coords=bool(vv.attrib)
|
||||
if coords:
|
||||
p_h=vv.attrib['points'].split(' ')
|
||||
c_t_in.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
break
|
||||
else:
|
||||
pass
|
||||
|
||||
|
||||
if vv.tag==link+'Point':
|
||||
c_t_in.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
|
||||
elif vv.tag!=link+'Point' and sumi>=1:
|
||||
break
|
||||
co_table.append(np.array(c_t_in))
|
||||
co_table_text.append(' ')
|
||||
|
||||
elif tag.endswith('}NoiseRegion') or tag.endswith('}noiseregion'):
|
||||
for nn in root1.iter(tag):
|
||||
c_t_in=[]
|
||||
sumi=0
|
||||
for vv in nn.iter():
|
||||
# check the format of coords
|
||||
if vv.tag==link+'Coords':
|
||||
coords=bool(vv.attrib)
|
||||
if coords:
|
||||
p_h=vv.attrib['points'].split(' ')
|
||||
c_t_in.append( np.array( [ [ int(x.split(',')[0]) , int(x.split(',')[1]) ] for x in p_h] ) )
|
||||
break
|
||||
else:
|
||||
pass
|
||||
|
||||
|
||||
if vv.tag==link+'Point':
|
||||
c_t_in.append([ int(float(vv.attrib['x'])) , int(float(vv.attrib['y'])) ])
|
||||
sumi+=1
|
||||
|
||||
elif vv.tag!=link+'Point' and sumi>=1:
|
||||
break
|
||||
co_noise.append(np.array(c_t_in))
|
||||
co_noise_text.append(' ')
|
||||
|
||||
img = np.zeros( (y_len,x_len,3) )
|
||||
img_poly=cv2.fillPoly(img, pts =co_text_paragraph, color=(1,1,1))
|
||||
|
||||
img_poly=cv2.fillPoly(img, pts =co_text_heading, color=(2,2,2))
|
||||
img_poly=cv2.fillPoly(img, pts =co_text_header, color=(2,2,2))
|
||||
img_poly=cv2.fillPoly(img, pts =co_text_marginalia, color=(3,3,3))
|
||||
img_poly=cv2.fillPoly(img, pts =co_img, color=(4,4,4))
|
||||
img_poly=cv2.fillPoly(img, pts =co_sep, color=(5,5,5))
|
||||
|
||||
return tree1, root1, bb_coord_printspace, id_paragraph, id_header+id_heading, co_text_paragraph, co_text_header+co_text_heading,\
|
||||
tot_region_ref,x_len, y_len,index_tot_regions, img_poly
|
||||
|
||||
def return_indexes_of_contours_loctaed_inside_another_list_of_contours(self, contours, contours_loc, cx_main_loc, cy_main_loc, indexes_loc):
|
||||
indexes_of_located_cont = []
|
||||
center_x_coordinates_of_located = []
|
||||
center_y_coordinates_of_located = []
|
||||
#M_main_tot = [cv2.moments(contours_loc[j])
|
||||
#for j in range(len(contours_loc))]
|
||||
#cx_main_loc = [(M_main_tot[j]["m10"] / (M_main_tot[j]["m00"] + 1e-32)) for j in range(len(M_main_tot))]
|
||||
#cy_main_loc = [(M_main_tot[j]["m01"] / (M_main_tot[j]["m00"] + 1e-32)) for j in range(len(M_main_tot))]
|
||||
|
||||
for ij in range(len(contours)):
|
||||
results = [cv2.pointPolygonTest(contours[ij], (cx_main_loc[ind], cy_main_loc[ind]), False)
|
||||
for ind in range(len(cy_main_loc)) ]
|
||||
results = np.array(results)
|
||||
indexes_in = np.where((results == 0) | (results == 1))
|
||||
indexes = indexes_loc[indexes_in]# [(results == 0) | (results == 1)]#np.where((results == 0) | (results == 1))
|
||||
|
||||
indexes_of_located_cont.append(indexes)
|
||||
center_x_coordinates_of_located.append(np.array(cx_main_loc)[indexes_in] )
|
||||
center_y_coordinates_of_located.append(np.array(cy_main_loc)[indexes_in] )
|
||||
|
||||
return indexes_of_located_cont, center_x_coordinates_of_located, center_y_coordinates_of_located
|
||||
|
||||
def do_order_of_regions_with_model(self, contours_only_text_parent, contours_only_text_parent_h, text_regions_p):
|
||||
height1 =672#448
|
||||
width1 = 448#224
|
||||
|
||||
height2 =672#448
|
||||
width2= 448#224
|
||||
|
||||
height3 =672#448
|
||||
width3 = 448#224
|
||||
|
||||
inference_bs = 3
|
||||
|
||||
ver_kernel = np.ones((5, 1), dtype=np.uint8)
|
||||
hor_kernel = np.ones((1, 5), dtype=np.uint8)
|
||||
|
||||
|
||||
min_cont_size_to_be_dilated = 10
|
||||
if len(contours_only_text_parent)>min_cont_size_to_be_dilated:
|
||||
cx_conts, cy_conts, x_min_conts, x_max_conts, y_min_conts, y_max_conts, _ = find_new_features_of_contours(contours_only_text_parent)
|
||||
args_cont_located = np.array(range(len(contours_only_text_parent)))
|
||||
|
||||
diff_y_conts = np.abs(y_max_conts[:]-y_min_conts)
|
||||
diff_x_conts = np.abs(x_max_conts[:]-x_min_conts)
|
||||
|
||||
mean_x = statistics.mean(diff_x_conts)
|
||||
median_x = statistics.median(diff_x_conts)
|
||||
|
||||
|
||||
diff_x_ratio= diff_x_conts/mean_x
|
||||
|
||||
args_cont_located_excluded = args_cont_located[diff_x_ratio>=1.3]
|
||||
args_cont_located_included = args_cont_located[diff_x_ratio<1.3]
|
||||
|
||||
contours_only_text_parent_excluded = [contours_only_text_parent[ind] for ind in range(len(contours_only_text_parent)) if diff_x_ratio[ind]>=1.3]#contours_only_text_parent[diff_x_ratio>=1.3]
|
||||
contours_only_text_parent_included = [contours_only_text_parent[ind] for ind in range(len(contours_only_text_parent)) if diff_x_ratio[ind]<1.3]#contours_only_text_parent[diff_x_ratio<1.3]
|
||||
|
||||
|
||||
cx_conts_excluded = [cx_conts[ind] for ind in range(len(cx_conts)) if diff_x_ratio[ind]>=1.3]#cx_conts[diff_x_ratio>=1.3]
|
||||
cx_conts_included = [cx_conts[ind] for ind in range(len(cx_conts)) if diff_x_ratio[ind]<1.3]#cx_conts[diff_x_ratio<1.3]
|
||||
|
||||
cy_conts_excluded = [cy_conts[ind] for ind in range(len(cy_conts)) if diff_x_ratio[ind]>=1.3]#cy_conts[diff_x_ratio>=1.3]
|
||||
cy_conts_included = [cy_conts[ind] for ind in range(len(cy_conts)) if diff_x_ratio[ind]<1.3]#cy_conts[diff_x_ratio<1.3]
|
||||
|
||||
#print(diff_x_ratio, 'ratio')
|
||||
text_regions_p = text_regions_p.astype('uint8')
|
||||
|
||||
if len(contours_only_text_parent_excluded)>0:
|
||||
textregion_par = np.zeros((text_regions_p.shape[0], text_regions_p.shape[1])).astype('uint8')
|
||||
textregion_par = cv2.fillPoly(textregion_par, pts=contours_only_text_parent_included, color=(1,1))
|
||||
else:
|
||||
textregion_par = (text_regions_p[:,:]==1)*1
|
||||
textregion_par = textregion_par.astype('uint8')
|
||||
|
||||
text_regions_p_textregions_dilated = cv2.erode(textregion_par , hor_kernel, iterations=2)
|
||||
text_regions_p_textregions_dilated = cv2.dilate(text_regions_p_textregions_dilated , ver_kernel, iterations=4)
|
||||
text_regions_p_textregions_dilated = cv2.erode(text_regions_p_textregions_dilated , hor_kernel, iterations=1)
|
||||
text_regions_p_textregions_dilated = cv2.dilate(text_regions_p_textregions_dilated , ver_kernel, iterations=5)
|
||||
text_regions_p_textregions_dilated[text_regions_p[:,:]>1] = 0
|
||||
|
||||
|
||||
contours_only_dilated, hir_on_text_dilated = return_contours_of_image(text_regions_p_textregions_dilated)
|
||||
contours_only_dilated = return_parent_contours(contours_only_dilated, hir_on_text_dilated)
|
||||
|
||||
indexes_of_located_cont, center_x_coordinates_of_located, center_y_coordinates_of_located = self.return_indexes_of_contours_loctaed_inside_another_list_of_contours(contours_only_dilated, contours_only_text_parent_included, cx_conts_included, cy_conts_included, args_cont_located_included)
|
||||
|
||||
|
||||
if len(args_cont_located_excluded)>0:
|
||||
for ind in args_cont_located_excluded:
|
||||
indexes_of_located_cont.append(np.array([ind]))
|
||||
contours_only_dilated.append(contours_only_text_parent[ind])
|
||||
center_y_coordinates_of_located.append(0)
|
||||
|
||||
array_list = [np.array([elem]) if isinstance(elem, int) else elem for elem in indexes_of_located_cont]
|
||||
flattened_array = np.concatenate([arr.ravel() for arr in array_list])
|
||||
#print(len( np.unique(flattened_array)), 'indexes_of_located_cont uniques')
|
||||
|
||||
missing_textregions = list( set(np.array(range(len(contours_only_text_parent))) ) - set(np.unique(flattened_array)) )
|
||||
#print(missing_textregions, 'missing_textregions')
|
||||
|
||||
for ind in missing_textregions:
|
||||
indexes_of_located_cont.append(np.array([ind]))
|
||||
contours_only_dilated.append(contours_only_text_parent[ind])
|
||||
center_y_coordinates_of_located.append(0)
|
||||
|
||||
|
||||
if contours_only_text_parent_h:
|
||||
for vi in range(len(contours_only_text_parent_h)):
|
||||
indexes_of_located_cont.append(int(vi+len(contours_only_text_parent)))
|
||||
|
||||
array_list = [np.array([elem]) if isinstance(elem, int) else elem for elem in indexes_of_located_cont]
|
||||
flattened_array = np.concatenate([arr.ravel() for arr in array_list])
|
||||
|
||||
y_len = text_regions_p.shape[0]
|
||||
x_len = text_regions_p.shape[1]
|
||||
|
||||
img_poly = np.zeros((y_len,x_len), dtype='uint8')
|
||||
###img_poly[text_regions_p[:,:]==1] = 1
|
||||
###img_poly[text_regions_p[:,:]==2] = 2
|
||||
###img_poly[text_regions_p[:,:]==3] = 4
|
||||
###img_poly[text_regions_p[:,:]==6] = 5
|
||||
|
||||
##img_poly[text_regions_p[:,:]==1] = 1
|
||||
##img_poly[text_regions_p[:,:]==2] = 2
|
||||
##img_poly[text_regions_p[:,:]==3] = 3
|
||||
##img_poly[text_regions_p[:,:]==4] = 4
|
||||
##img_poly[text_regions_p[:,:]==5] = 5
|
||||
|
||||
img_poly = np.copy(text_regions_p)
|
||||
|
||||
img_header_and_sep = np.zeros((y_len,x_len), dtype='uint8')
|
||||
if contours_only_text_parent_h:
|
||||
_, cy_main, x_min_main, x_max_main, y_min_main, y_max_main, _ = find_new_features_of_contours(
|
||||
contours_only_text_parent_h)
|
||||
for j in range(len(cy_main)):
|
||||
img_header_and_sep[int(y_max_main[j]):int(y_max_main[j])+12,
|
||||
int(x_min_main[j]):int(x_max_main[j])] = 1
|
||||
co_text_all_org = contours_only_text_parent + contours_only_text_parent_h
|
||||
if len(contours_only_text_parent)>min_cont_size_to_be_dilated:
|
||||
co_text_all = contours_only_dilated + contours_only_text_parent_h
|
||||
else:
|
||||
co_text_all = contours_only_text_parent + contours_only_text_parent_h
|
||||
else:
|
||||
co_text_all_org = contours_only_text_parent
|
||||
if len(contours_only_text_parent)>min_cont_size_to_be_dilated:
|
||||
co_text_all = contours_only_dilated
|
||||
else:
|
||||
co_text_all = contours_only_text_parent
|
||||
|
||||
if not len(co_text_all):
|
||||
return [], []
|
||||
|
||||
labels_con = np.zeros((int(y_len /6.), int(x_len/6.), len(co_text_all)), dtype=bool)
|
||||
|
||||
co_text_all = [(i/6).astype(int) for i in co_text_all]
|
||||
for i in range(len(co_text_all)):
|
||||
img = labels_con[:,:,i].astype(np.uint8)
|
||||
|
||||
#img = cv2.resize(img, (int(img.shape[1]/6), int(img.shape[0]/6)), interpolation=cv2.INTER_NEAREST)
|
||||
|
||||
cv2.fillPoly(img, pts=[co_text_all[i]], color=(1,))
|
||||
labels_con[:,:,i] = img
|
||||
|
||||
|
||||
labels_con = resize_image(labels_con.astype(np.uint8), height1, width1).astype(bool)
|
||||
img_header_and_sep = resize_image(img_header_and_sep, height1, width1)
|
||||
img_poly = resize_image(img_poly, height3, width3)
|
||||
|
||||
|
||||
|
||||
input_1 = np.zeros((inference_bs, height1, width1, 3))
|
||||
ordered = [list(range(len(co_text_all)))]
|
||||
index_update = 0
|
||||
#print(labels_con.shape[2],"number of regions for reading order")
|
||||
while index_update>=0:
|
||||
ij_list = ordered.pop(index_update)
|
||||
i = ij_list.pop(0)
|
||||
|
||||
ante_list = []
|
||||
post_list = []
|
||||
tot_counter = 0
|
||||
batch = []
|
||||
for j in ij_list:
|
||||
img1 = labels_con[:,:,i].astype(float)
|
||||
img2 = labels_con[:,:,j].astype(float)
|
||||
img1[img_poly==5] = 2
|
||||
img2[img_poly==5] = 2
|
||||
img1[img_header_and_sep==1] = 3
|
||||
img2[img_header_and_sep==1] = 3
|
||||
|
||||
input_1[len(batch), :, :, 0] = img1 / 3.
|
||||
input_1[len(batch), :, :, 2] = img2 / 3.
|
||||
input_1[len(batch), :, :, 1] = img_poly / 5.
|
||||
|
||||
tot_counter += 1
|
||||
batch.append(j)
|
||||
if tot_counter % inference_bs == 0 or tot_counter == len(ij_list):
|
||||
y_pr = self.model_zoo.get('reading_order', Model).predict(input_1 , verbose='0')
|
||||
for jb, j in enumerate(batch):
|
||||
if y_pr[jb][0]>=0.5:
|
||||
post_list.append(j)
|
||||
else:
|
||||
ante_list.append(j)
|
||||
batch = []
|
||||
|
||||
if len(ante_list):
|
||||
ordered.insert(index_update, ante_list)
|
||||
index_update += 1
|
||||
ordered.insert(index_update, [i])
|
||||
if len(post_list):
|
||||
ordered.insert(index_update + 1, post_list)
|
||||
|
||||
index_update = -1
|
||||
for index_next, ij_list in enumerate(ordered):
|
||||
if len(ij_list) > 1:
|
||||
index_update = index_next
|
||||
break
|
||||
|
||||
ordered = [i[0] for i in ordered]
|
||||
|
||||
##id_all_text = np.array(id_all_text)[index_sort]
|
||||
|
||||
|
||||
if len(contours_only_text_parent)>min_cont_size_to_be_dilated:
|
||||
org_contours_indexes = []
|
||||
for ind in range(len(ordered)):
|
||||
region_with_curr_order = ordered[ind]
|
||||
if region_with_curr_order < len(contours_only_dilated):
|
||||
if np.isscalar(indexes_of_located_cont[region_with_curr_order]):
|
||||
org_contours_indexes = org_contours_indexes + [indexes_of_located_cont[region_with_curr_order]]
|
||||
else:
|
||||
arg_sort_located_cont = np.argsort(center_y_coordinates_of_located[region_with_curr_order])
|
||||
org_contours_indexes = org_contours_indexes + list(np.array(indexes_of_located_cont[region_with_curr_order])[arg_sort_located_cont]) ##org_contours_indexes + list (
|
||||
else:
|
||||
org_contours_indexes = org_contours_indexes + [indexes_of_located_cont[region_with_curr_order]]
|
||||
|
||||
region_ids = ['region_%04d' % i for i in range(len(co_text_all_org))]
|
||||
return org_contours_indexes, region_ids
|
||||
else:
|
||||
region_ids = ['region_%04d' % i for i in range(len(co_text_all_org))]
|
||||
return ordered, region_ids
|
||||
|
||||
|
||||
|
||||
|
||||
def run(self,
|
||||
overwrite: bool = False,
|
||||
xml_filename: Optional[str] = None,
|
||||
dir_in: Optional[str] = None,
|
||||
dir_out: Optional[str] = None,
|
||||
):
|
||||
"""
|
||||
Get image and scales, then extract the page of scanned image
|
||||
"""
|
||||
self.logger.debug("enter run")
|
||||
t0_tot = time.time()
|
||||
|
||||
if dir_in:
|
||||
ls_xmls = [os.path.join(dir_in, xml_filename)
|
||||
for xml_filename in filter(is_xml_filename,
|
||||
os.listdir(dir_in))]
|
||||
elif xml_filename:
|
||||
ls_xmls = [xml_filename]
|
||||
else:
|
||||
raise ValueError("run requires either a single image filename or a directory")
|
||||
|
||||
for xml_filename in ls_xmls:
|
||||
self.logger.info(xml_filename)
|
||||
t0 = time.time()
|
||||
|
||||
file_name = Path(xml_filename).stem
|
||||
(tree_xml, root_xml, bb_coord_printspace, id_paragraph, id_header,
|
||||
co_text_paragraph, co_text_header, tot_region_ref,
|
||||
x_len, y_len, index_tot_regions, img_poly) = self.read_xml(xml_filename)
|
||||
|
||||
id_all_text = id_paragraph + id_header
|
||||
|
||||
order_text_new, id_of_texts_tot = self.do_order_of_regions_with_model(co_text_paragraph, co_text_header, img_poly[:,:,0])
|
||||
|
||||
id_all_text = np.array(id_all_text)[order_text_new]
|
||||
|
||||
alltags=[elem.tag for elem in root_xml.iter()]
|
||||
|
||||
|
||||
|
||||
link=alltags[0].split('}')[0]+'}'
|
||||
name_space = alltags[0].split('}')[0]
|
||||
name_space = name_space.split('{')[1]
|
||||
|
||||
page_element = root_xml.find(link+'Page')
|
||||
|
||||
|
||||
old_ro = root_xml.find(".//{*}ReadingOrder")
|
||||
|
||||
if old_ro is not None:
|
||||
page_element.remove(old_ro)
|
||||
|
||||
#print(old_ro, 'old_ro')
|
||||
ro_subelement = ET.Element('ReadingOrder')
|
||||
|
||||
ro_subelement2 = ET.SubElement(ro_subelement, 'OrderedGroup')
|
||||
ro_subelement2.set('id', "ro357564684568544579089")
|
||||
|
||||
for index, id_text in enumerate(id_all_text):
|
||||
new_element_2 = ET.SubElement(ro_subelement2, 'RegionRefIndexed')
|
||||
new_element_2.set('regionRef', id_all_text[index])
|
||||
new_element_2.set('index', str(index))
|
||||
|
||||
if (link+'PrintSpace' in alltags) or (link+'Border' in alltags):
|
||||
page_element.insert(1, ro_subelement)
|
||||
else:
|
||||
page_element.insert(0, ro_subelement)
|
||||
|
||||
alltags=[elem.tag for elem in root_xml.iter()]
|
||||
|
||||
ET.register_namespace("",name_space)
|
||||
assert dir_out
|
||||
tree_xml.write(os.path.join(dir_out, file_name+'.xml'),
|
||||
xml_declaration=True,
|
||||
method='xml',
|
||||
encoding="utf-8",
|
||||
default_namespace=None)
|
||||
|
||||
#sys.exit()
|
||||
|
||||
|
|
@ -1,4 +0,0 @@
|
|||
__all__ = [
|
||||
'EynollahModelZoo',
|
||||
]
|
||||
from .model_zoo import EynollahModelZoo
|
||||
|
|
@ -1,252 +0,0 @@
|
|||
from .specs import EynollahModelSpec, EynollahModelSpecSet
|
||||
|
||||
# NOTE: This needs to change whenever models/versions change
|
||||
ZENODO = "https://zenodo.org/records/17295988/files"
|
||||
MODELS_VERSION = "v0_7_0"
|
||||
|
||||
def dist_url(dist_name: str="layout") -> str:
|
||||
return f'{ZENODO}/models_{dist_name}_{MODELS_VERSION}.zip'
|
||||
|
||||
DEFAULT_MODEL_SPECS = EynollahModelSpecSet([
|
||||
|
||||
EynollahModelSpec(
|
||||
category="enhancement",
|
||||
variant='',
|
||||
filename="models_eynollah/eynollah-enhancement_20210425",
|
||||
dist_url=dist_url(),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="binarization",
|
||||
variant='hybrid',
|
||||
filename="models_eynollah/eynollah-binarization-hybrid_20230504/model_bin_hybrid_trans_cnn_sbb_ens",
|
||||
dist_url=dist_url(),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="binarization",
|
||||
variant='20210309',
|
||||
filename="models_eynollah/eynollah-binarization_20210309",
|
||||
dist_url=dist_url("extra"),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="binarization",
|
||||
variant='',
|
||||
filename="models_eynollah/eynollah-binarization_20210425",
|
||||
dist_url=dist_url("extra"),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="col_classifier",
|
||||
variant='',
|
||||
filename="models_eynollah/eynollah-column-classifier_20210425",
|
||||
dist_url=dist_url(),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="page",
|
||||
variant='',
|
||||
filename="models_eynollah/model_eynollah_page_extraction_20250915",
|
||||
dist_url=dist_url(),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="region",
|
||||
variant='',
|
||||
filename="models_eynollah/eynollah-main-regions-ensembled_20210425",
|
||||
dist_url=dist_url(),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="extract_images",
|
||||
variant='',
|
||||
filename="models_eynollah/eynollah-main-regions_20231127_672_org_ens_11_13_16_17_18",
|
||||
dist_url=dist_url(),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="region",
|
||||
variant='',
|
||||
filename="models_eynollah/eynollah-main-regions_20220314",
|
||||
dist_url=dist_url(),
|
||||
help="early layout",
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="region_p2",
|
||||
variant='non-light',
|
||||
filename="models_eynollah/eynollah-main-regions-aug-rotation_20210425",
|
||||
dist_url=dist_url('extra'),
|
||||
help="early layout, non-light, 2nd part",
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="region_1_2",
|
||||
variant='',
|
||||
#filename="models_eynollah/modelens_12sp_elay_0_3_4__3_6_n",
|
||||
#filename="models_eynollah/modelens_earlylayout_12spaltige_2_3_5_6_7_8",
|
||||
#filename="models_eynollah/modelens_early12_sp_2_3_5_6_7_8_9_10_12_14_15_16_18",
|
||||
#filename="models_eynollah/modelens_1_2_4_5_early_lay_1_2_spaltige",
|
||||
#filename="models_eynollah/model_3_eraly_layout_no_patches_1_2_spaltige",
|
||||
filename="models_eynollah/modelens_e_l_all_sp_0_1_2_3_4_171024",
|
||||
dist_url=dist_url("layout"),
|
||||
help="early layout, light, 1-or-2-column",
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="region_fl_np",
|
||||
variant='',
|
||||
#'filename="models_eynollah/modelens_full_lay_1_3_031124",
|
||||
#'filename="models_eynollah/modelens_full_lay_13__3_19_241024",
|
||||
#'filename="models_eynollah/model_full_lay_13_241024",
|
||||
#'filename="models_eynollah/modelens_full_lay_13_17_231024",
|
||||
#'filename="models_eynollah/modelens_full_lay_1_2_221024",
|
||||
#'filename="models_eynollah/eynollah-full-regions-1column_20210425",
|
||||
filename="models_eynollah/modelens_full_lay_1__4_3_091124",
|
||||
dist_url=dist_url(),
|
||||
help="full layout / no patches",
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
# FIXME: Why is region_fl and region_fl_np the same model?
|
||||
EynollahModelSpec(
|
||||
category="region_fl",
|
||||
variant='',
|
||||
# filename="models_eynollah/eynollah-full-regions-3+column_20210425",
|
||||
# filename="models_eynollah/model_2_full_layout_new_trans",
|
||||
# filename="models_eynollah/modelens_full_lay_1_3_031124",
|
||||
# filename="models_eynollah/modelens_full_lay_13__3_19_241024",
|
||||
# filename="models_eynollah/model_full_lay_13_241024",
|
||||
# filename="models_eynollah/modelens_full_lay_13_17_231024",
|
||||
# filename="models_eynollah/modelens_full_lay_1_2_221024",
|
||||
# filename="models_eynollah/modelens_full_layout_24_till_28",
|
||||
# filename="models_eynollah/model_2_full_layout_new_trans",
|
||||
filename="models_eynollah/modelens_full_lay_1__4_3_091124",
|
||||
dist_url=dist_url(),
|
||||
help="full layout / with patches",
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="reading_order",
|
||||
variant='',
|
||||
#filename="models_eynollah/model_mb_ro_aug_ens_11",
|
||||
#filename="models_eynollah/model_step_3200000_mb_ro",
|
||||
#filename="models_eynollah/model_ens_reading_order_machine_based",
|
||||
#filename="models_eynollah/model_mb_ro_aug_ens_8",
|
||||
#filename="models_eynollah/model_ens_reading_order_machine_based",
|
||||
filename="models_eynollah/model_eynollah_reading_order_20250824",
|
||||
dist_url=dist_url(),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="textline",
|
||||
variant='non-light',
|
||||
#filename="models_eynollah/modelens_textline_1_4_16092024",
|
||||
#filename="models_eynollah/model_textline_ens_3_4_5_6_artificial",
|
||||
#filename="models_eynollah/modelens_textline_1_3_4_20240915",
|
||||
#filename="models_eynollah/model_textline_ens_3_4_5_6_artificial",
|
||||
#filename="models_eynollah/modelens_textline_9_12_13_14_15",
|
||||
#filename="models_eynollah/eynollah-textline_20210425",
|
||||
filename="models_eynollah/modelens_textline_0_1__2_4_16092024",
|
||||
dist_url=dist_url('extra'),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="textline",
|
||||
variant='',
|
||||
#filename="models_eynollah/eynollah-textline_light_20210425",
|
||||
filename="models_eynollah/modelens_textline_0_1__2_4_16092024",
|
||||
dist_url=dist_url(),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="table",
|
||||
variant='non-light',
|
||||
filename="models_eynollah/eynollah-tables_20210319",
|
||||
dist_url=dist_url('extra'),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="table",
|
||||
variant='',
|
||||
filename="models_eynollah/modelens_table_0t4_201124",
|
||||
dist_url=dist_url(),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="ocr",
|
||||
variant='',
|
||||
filename="models_eynollah/model_eynollah_ocr_cnnrnn_20250930",
|
||||
dist_url=dist_url("ocr"),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="ocr",
|
||||
variant='degraded',
|
||||
filename="models_eynollah/model_eynollah_ocr_cnnrnn__degraded_20250805/",
|
||||
help="slightly better at degraded Fraktur",
|
||||
dist_url=dist_url("ocr"),
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="num_to_char",
|
||||
variant='',
|
||||
filename="characters_org.txt",
|
||||
dist_url=dist_url("ocr"),
|
||||
type='decoder',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="characters",
|
||||
variant='',
|
||||
filename="characters_org.txt",
|
||||
dist_url=dist_url("ocr"),
|
||||
type='List[str]',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="ocr",
|
||||
variant='tr',
|
||||
filename="models_eynollah/model_eynollah_ocr_trocr_20250919",
|
||||
dist_url=dist_url("ocr"),
|
||||
help='much slower transformer-based',
|
||||
type='Keras',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="trocr_processor",
|
||||
variant='',
|
||||
filename="models_eynollah/model_eynollah_ocr_trocr_20250919",
|
||||
dist_url=dist_url("ocr"),
|
||||
type='TrOCRProcessor',
|
||||
),
|
||||
|
||||
EynollahModelSpec(
|
||||
category="trocr_processor",
|
||||
variant='htr',
|
||||
filename="models_eynollah/microsoft/trocr-base-handwritten",
|
||||
dist_url=dist_url("extra"),
|
||||
type='TrOCRProcessor',
|
||||
),
|
||||
|
||||
])
|
||||
|
|
@ -1,203 +0,0 @@
|
|||
import json
|
||||
import logging
|
||||
from copy import deepcopy
|
||||
from pathlib import Path
|
||||
from typing import Dict, List, Optional, Tuple, Type, Union
|
||||
|
||||
from ocrd_utils import tf_disable_interactive_logs
|
||||
tf_disable_interactive_logs()
|
||||
|
||||
from keras.layers import StringLookup
|
||||
from keras.models import Model as KerasModel
|
||||
from keras.models import load_model
|
||||
from tabulate import tabulate
|
||||
from ..patch_encoder import PatchEncoder, Patches
|
||||
from .specs import EynollahModelSpecSet
|
||||
from .default_specs import DEFAULT_MODEL_SPECS
|
||||
from .types import AnyModel, T
|
||||
|
||||
|
||||
class EynollahModelZoo:
|
||||
"""
|
||||
Wrapper class that handles storage and loading of models for all eynollah runners.
|
||||
"""
|
||||
|
||||
model_basedir: Path
|
||||
specs: EynollahModelSpecSet
|
||||
|
||||
def __init__(
|
||||
self,
|
||||
basedir: str,
|
||||
model_overrides: Optional[List[Tuple[str, str, str]]] = None,
|
||||
) -> None:
|
||||
self.model_basedir = Path(basedir)
|
||||
self.logger = logging.getLogger('eynollah.model_zoo')
|
||||
if not self.model_basedir.exists():
|
||||
self.logger.warning(f"Model basedir does not exist: {basedir}. Set eynollah --model-basedir to the correct directory.")
|
||||
self.specs = deepcopy(DEFAULT_MODEL_SPECS)
|
||||
self._overrides = []
|
||||
if model_overrides:
|
||||
self.override_models(*model_overrides)
|
||||
self._loaded: Dict[str, AnyModel] = {}
|
||||
|
||||
@property
|
||||
def model_overrides(self):
|
||||
return self._overrides
|
||||
|
||||
def override_models(
|
||||
self,
|
||||
*model_overrides: Tuple[str, str, str],
|
||||
):
|
||||
"""
|
||||
Override the default model versions
|
||||
"""
|
||||
for model_category, model_variant, model_filename in model_overrides:
|
||||
spec = self.specs.get(model_category, model_variant)
|
||||
self.logger.warning("Overriding filename for model spec %s to %s", spec, model_filename)
|
||||
self.specs.get(model_category, model_variant).filename = model_filename
|
||||
self._overrides += model_overrides
|
||||
|
||||
def model_path(
|
||||
self,
|
||||
model_category: str,
|
||||
model_variant: str = '',
|
||||
absolute: bool = True,
|
||||
) -> Path:
|
||||
"""
|
||||
Translate model_{type,variant} tuple into an absolute (or relative) Path
|
||||
"""
|
||||
spec = self.specs.get(model_category, model_variant)
|
||||
if spec.category in ('characters', 'num_to_char'):
|
||||
return self.model_path('ocr') / spec.filename
|
||||
if not Path(spec.filename).is_absolute() and absolute:
|
||||
model_path = Path(self.model_basedir).joinpath(spec.filename)
|
||||
else:
|
||||
model_path = Path(spec.filename)
|
||||
return model_path
|
||||
|
||||
def load_models(
|
||||
self,
|
||||
*all_load_args: Union[str, Tuple[str], Tuple[str, str], Tuple[str, str, str]],
|
||||
) -> Dict:
|
||||
"""
|
||||
Load all models by calling load_model and return a dictionary mapping model_category to loaded model
|
||||
"""
|
||||
ret = {}
|
||||
for load_args in all_load_args:
|
||||
if isinstance(load_args, str):
|
||||
ret[load_args] = self.load_model(load_args)
|
||||
else:
|
||||
ret[load_args[0]] = self.load_model(*load_args)
|
||||
return ret
|
||||
|
||||
def load_model(
|
||||
self,
|
||||
model_category: str,
|
||||
model_variant: str = '',
|
||||
model_path_override: Optional[str] = None,
|
||||
) -> AnyModel:
|
||||
"""
|
||||
Load any model
|
||||
"""
|
||||
if model_path_override:
|
||||
self.override_models((model_category, model_variant, model_path_override))
|
||||
model_path = self.model_path(model_category, model_variant)
|
||||
if model_path.suffix == '.h5' and Path(model_path.stem).exists():
|
||||
# prefer SavedModel over HDF5 format if it exists
|
||||
model_path = Path(model_path.stem)
|
||||
if model_category == 'ocr':
|
||||
model = self._load_ocr_model(variant=model_variant)
|
||||
elif model_category == 'num_to_char':
|
||||
model = self._load_num_to_char()
|
||||
elif model_category == 'characters':
|
||||
model = self._load_characters()
|
||||
elif model_category == 'trocr_processor':
|
||||
from transformers import TrOCRProcessor
|
||||
model = TrOCRProcessor.from_pretrained(model_path)
|
||||
else:
|
||||
try:
|
||||
model = load_model(model_path, compile=False)
|
||||
except Exception as e:
|
||||
self.logger.exception(e)
|
||||
model = load_model(
|
||||
model_path, compile=False, custom_objects={"PatchEncoder": PatchEncoder, "Patches": Patches}
|
||||
)
|
||||
self._loaded[model_category] = model
|
||||
return model # type: ignore
|
||||
|
||||
def get(self, model_category: str, model_type: Optional[Type[T]] = None) -> T:
|
||||
if model_category not in self._loaded:
|
||||
raise ValueError(f'Model "{model_category} not previously loaded with "load_model(..)"')
|
||||
ret = self._loaded[model_category]
|
||||
if model_type:
|
||||
assert isinstance(ret, model_type)
|
||||
return ret # type: ignore # FIXME: convince typing that we're returning generic type
|
||||
|
||||
def _load_ocr_model(self, variant: str) -> AnyModel:
|
||||
"""
|
||||
Load OCR model
|
||||
"""
|
||||
ocr_model_dir = self.model_path('ocr', variant)
|
||||
if variant == 'tr':
|
||||
from transformers import VisionEncoderDecoderModel
|
||||
ret = VisionEncoderDecoderModel.from_pretrained(ocr_model_dir)
|
||||
assert isinstance(ret, VisionEncoderDecoderModel)
|
||||
return ret
|
||||
else:
|
||||
ocr_model = load_model(ocr_model_dir, compile=False)
|
||||
assert isinstance(ocr_model, KerasModel)
|
||||
return KerasModel(
|
||||
ocr_model.get_layer(name="image").input, # type: ignore
|
||||
ocr_model.get_layer(name="dense2").output, # type: ignore
|
||||
)
|
||||
|
||||
def _load_characters(self) -> List[str]:
|
||||
"""
|
||||
Load encoding for OCR
|
||||
"""
|
||||
with open(self.model_path('num_to_char'), "r") as config_file:
|
||||
return json.load(config_file)
|
||||
|
||||
def _load_num_to_char(self) -> StringLookup:
|
||||
"""
|
||||
Load decoder for OCR
|
||||
"""
|
||||
characters = self._load_characters()
|
||||
# Mapping characters to integers.
|
||||
char_to_num = StringLookup(vocabulary=characters, mask_token=None)
|
||||
# Mapping integers back to original characters.
|
||||
return StringLookup(vocabulary=char_to_num.get_vocabulary(), mask_token=None, invert=True)
|
||||
|
||||
def __str__(self):
|
||||
return tabulate(
|
||||
[
|
||||
[
|
||||
spec.type,
|
||||
spec.category,
|
||||
spec.variant,
|
||||
spec.help,
|
||||
f'Yes, at {self.model_path(spec.category, spec.variant)}'
|
||||
if self.model_path(spec.category, spec.variant).exists()
|
||||
else f'No, download {spec.dist_url}',
|
||||
# self.model_path(spec.category, spec.variant),
|
||||
]
|
||||
for spec in sorted(self.specs.specs, key=lambda x: x.dist_url)
|
||||
],
|
||||
headers=[
|
||||
'Type',
|
||||
'Category',
|
||||
'Variant',
|
||||
'Help',
|
||||
'Used in',
|
||||
'Installed',
|
||||
],
|
||||
tablefmt='github',
|
||||
)
|
||||
|
||||
def shutdown(self):
|
||||
"""
|
||||
Ensure that a loaded models is not referenced by ``self._loaded`` anymore
|
||||
"""
|
||||
if hasattr(self, '_loaded') and getattr(self, '_loaded'):
|
||||
for needle in list(self._loaded.keys()):
|
||||
del self._loaded[needle]
|
||||
|
|
@ -1,52 +0,0 @@
|
|||
from dataclasses import dataclass
|
||||
from typing import Dict, List, Set, Tuple
|
||||
|
||||
|
||||
@dataclass
|
||||
class EynollahModelSpec():
|
||||
"""
|
||||
Describing a single model abstractly.
|
||||
"""
|
||||
category: str
|
||||
# Relative filename to the models_eynollah directory in the dists
|
||||
filename: str
|
||||
# URL to the smallest model distribution containing this model (link to Zenodo)
|
||||
dist_url: str
|
||||
type: str
|
||||
variant: str = ''
|
||||
help: str = ''
|
||||
|
||||
class EynollahModelSpecSet():
|
||||
"""
|
||||
List of all used models for eynollah.
|
||||
"""
|
||||
specs: List[EynollahModelSpec]
|
||||
|
||||
def __init__(self, specs: List[EynollahModelSpec]) -> None:
|
||||
self.specs = sorted(specs, key=lambda x: x.category + '0' + x.variant)
|
||||
self.categories: Set[str] = set([spec.category for spec in self.specs])
|
||||
self.variants: Dict[str, Set[str]] = {
|
||||
spec.category: set([x.variant for x in self.specs if x.category == spec.category])
|
||||
for spec in self.specs
|
||||
}
|
||||
self._index_category_variant: Dict[Tuple[str, str], EynollahModelSpec] = {
|
||||
(spec.category, spec.variant): spec
|
||||
for spec in self.specs
|
||||
}
|
||||
|
||||
def asdict(self) -> Dict[str, Dict[str, str]]:
|
||||
return {
|
||||
spec.category: {
|
||||
spec.variant: spec.filename
|
||||
}
|
||||
for spec in self.specs
|
||||
}
|
||||
|
||||
def get(self, category: str, variant: str) -> EynollahModelSpec:
|
||||
if category not in self.categories:
|
||||
raise ValueError(f"Unknown category '{category}', must be one of {self.categories}")
|
||||
if variant not in self.variants[category]:
|
||||
raise ValueError(f"Unknown variant {variant} for {category}. Known variants: {self.variants[category]}")
|
||||
return self._index_category_variant[(category, variant)]
|
||||
|
||||
|
||||
|
|
@ -1,7 +0,0 @@
|
|||
from typing import TypeVar
|
||||
|
||||
# NOTE: Creating an actual union type requires loading transformers which is expensive and error-prone
|
||||
# from transformers import TrOCRProcessor, VisionEncoderDecoderModel
|
||||
# AnyModel = Union[VisionEncoderDecoderModel, TrOCRProcessor, KerasModel, List]
|
||||
AnyModel = object
|
||||
T = TypeVar('T')
|
||||
|
|
@ -1,5 +1,5 @@
|
|||
{
|
||||
"version": "0.7.0",
|
||||
"version": "0.4.0",
|
||||
"git_url": "https://github.com/qurator-spk/eynollah",
|
||||
"dockerhub": "ocrd/eynollah",
|
||||
"tools": {
|
||||
|
|
@ -29,6 +29,16 @@
|
|||
"type": "boolean",
|
||||
"default": true,
|
||||
"description": "Try to detect all element subtypes, including drop-caps and headings"
|
||||
},
|
||||
"light_version": {
|
||||
"type": "boolean",
|
||||
"default": true,
|
||||
"description": "Try to detect all element subtypes in light version (faster+simpler method for main region detection and deskewing)"
|
||||
},
|
||||
"textline_light": {
|
||||
"type": "boolean",
|
||||
"default": true,
|
||||
"description": "Light version need textline light"
|
||||
},
|
||||
"tables": {
|
||||
"type": "boolean",
|
||||
|
|
@ -55,6 +65,11 @@
|
|||
"default": false,
|
||||
"description": "if this parameter set to true, this tool would check that input image need resizing and enhancement or not."
|
||||
},
|
||||
"textline_light": {
|
||||
"type": "boolean",
|
||||
"default": false,
|
||||
"description": "if this parameter set to true, this tool will try to return contoure of textlines instead of rectangle bounding box of textline with a faster method."
|
||||
},
|
||||
"right_to_left": {
|
||||
"type": "boolean",
|
||||
"default": false,
|
||||
|
|
@ -64,39 +79,16 @@
|
|||
"type": "boolean",
|
||||
"default": false,
|
||||
"description": "ignore the special role of headings during reading order detection"
|
||||
},
|
||||
"reading_order_machine_based": {
|
||||
"type": "boolean",
|
||||
"default": false,
|
||||
"description": "use data-driven (rather than rule-based) reading order detection"
|
||||
}
|
||||
},
|
||||
"resources": [
|
||||
{
|
||||
"url": "https://zenodo.org/records/17580627/files/models_all_v0_7_0.zip?download=1",
|
||||
"name": "models_layout_v0_7_0",
|
||||
"type": "archive",
|
||||
"size": 6119874002,
|
||||
"description": "Models for layout detection, reading order detection, textline detection, page extraction, column classification, table detection, binarization, image enhancement and OCR",
|
||||
"version_range": ">= v0.7.0"
|
||||
},
|
||||
{
|
||||
"url": "https://zenodo.org/records/17295988/files/models_layout_v0_6_0.tar.gz?download=1",
|
||||
"name": "models_layout_v0_6_0",
|
||||
"type": "archive",
|
||||
"path_in_archive": "models_layout_v0_6_0",
|
||||
"size": 3525684179,
|
||||
"description": "Models for layout detection, reading order detection, textline detection, page extraction, column classification, table detection, binarization, image enhancement and OCR",
|
||||
"version_range": ">= v0.5.0"
|
||||
},
|
||||
{
|
||||
"description": "models for eynollah (TensorFlow SavedModel format)",
|
||||
"url": "https://github.com/qurator-spk/eynollah/releases/download/v0.3.1/models_eynollah.tar.gz",
|
||||
"name": "default",
|
||||
"size": 1894627041,
|
||||
"type": "archive",
|
||||
"path_in_archive": "models_eynollah",
|
||||
"version_range": ">= v0.3.0, < v0.5.0"
|
||||
"path_in_archive": "models_eynollah"
|
||||
}
|
||||
]
|
||||
},
|
||||
|
|
|
|||
|
|
@ -1,6 +1,3 @@
|
|||
# NOTE: For predictable order of imports of torch/shapely/tensorflow
|
||||
# this must be the first import of the CLI!
|
||||
from .eynollah_imports import imported_libs
|
||||
from .processor import EynollahProcessor
|
||||
from click import command
|
||||
from ocrd.decorators import ocrd_cli_options, ocrd_cli_wrap_processor
|
||||
|
|
|
|||
|
|
@ -1,8 +1,6 @@
|
|||
from functools import cached_property
|
||||
from typing import Optional
|
||||
|
||||
from PIL import Image
|
||||
from frozendict import frozendict
|
||||
import numpy as np
|
||||
import cv2
|
||||
from click import command
|
||||
|
|
@ -11,8 +9,6 @@ from ocrd import Processor, OcrdPageResult, OcrdPageResultImage
|
|||
from ocrd_models.ocrd_page import OcrdPage, AlternativeImageType
|
||||
from ocrd.decorators import ocrd_cli_options, ocrd_cli_wrap_processor
|
||||
|
||||
from eynollah.model_zoo.model_zoo import EynollahModelZoo
|
||||
|
||||
from .sbb_binarize import SbbBinarizer
|
||||
|
||||
|
||||
|
|
@ -29,7 +25,7 @@ class SbbBinarizeProcessor(Processor):
|
|||
# already employs GPU (without singleton process atm)
|
||||
max_workers = 1
|
||||
|
||||
@cached_property
|
||||
@property
|
||||
def executable(self):
|
||||
return 'ocrd-sbb-binarize'
|
||||
|
||||
|
|
@ -38,9 +34,8 @@ class SbbBinarizeProcessor(Processor):
|
|||
Set up the model prior to processing.
|
||||
"""
|
||||
# resolve relative path via OCR-D ResourceManager
|
||||
assert isinstance(self.parameter, frozendict)
|
||||
model_zoo = EynollahModelZoo(basedir=self.parameter['model'])
|
||||
self.binarizer = SbbBinarizer(model_zoo=model_zoo, logger=self.logger)
|
||||
model_path = self.resolve_resource(self.parameter['model'])
|
||||
self.binarizer = SbbBinarizer(model_dir=model_path, logger=self.logger)
|
||||
|
||||
def process_page_pcgts(self, *input_pcgts: Optional[OcrdPage], page_id: Optional[str] = None) -> OcrdPageResult:
|
||||
"""
|
||||
|
|
@ -103,7 +98,7 @@ class SbbBinarizeProcessor(Processor):
|
|||
line_image_bin = cv2pil(self.binarizer.run(image=pil2cv(line_image), use_patches=True))
|
||||
# update PAGE (reference the image file):
|
||||
line_image_ref = AlternativeImageType(comments=line_xywh['features'] + ',binarized')
|
||||
line.add_AlternativeImage(line_image_ref)
|
||||
line.add_AlternativeImage(region_image_ref)
|
||||
result.images.append(OcrdPageResultImage(line_image_bin, line.id + '.IMG-BIN', line_image_ref))
|
||||
|
||||
return result
|
||||
|
|
|
|||
|
|
@ -1,52 +0,0 @@
|
|||
from keras import layers
|
||||
import tensorflow as tf
|
||||
|
||||
projection_dim = 64
|
||||
patch_size = 1
|
||||
num_patches =21*21#14*14#28*28#14*14#28*28
|
||||
|
||||
class PatchEncoder(layers.Layer):
|
||||
|
||||
def __init__(self):
|
||||
super().__init__()
|
||||
self.projection = layers.Dense(units=projection_dim)
|
||||
self.position_embedding = layers.Embedding(input_dim=num_patches, output_dim=projection_dim)
|
||||
|
||||
def call(self, patch):
|
||||
positions = tf.range(start=0, limit=num_patches, delta=1)
|
||||
encoded = self.projection(patch) + self.position_embedding(positions)
|
||||
return encoded
|
||||
|
||||
def get_config(self):
|
||||
config = super().get_config().copy()
|
||||
config.update({
|
||||
'num_patches': num_patches,
|
||||
'projection': self.projection,
|
||||
'position_embedding': self.position_embedding,
|
||||
})
|
||||
return config
|
||||
|
||||
class Patches(layers.Layer):
|
||||
def __init__(self, **kwargs):
|
||||
super(Patches, self).__init__()
|
||||
self.patch_size = patch_size
|
||||
|
||||
def call(self, images):
|
||||
batch_size = tf.shape(images)[0]
|
||||
patches = tf.image.extract_patches(
|
||||
images=images,
|
||||
sizes=[1, self.patch_size, self.patch_size, 1],
|
||||
strides=[1, self.patch_size, self.patch_size, 1],
|
||||
rates=[1, 1, 1, 1],
|
||||
padding="VALID",
|
||||
)
|
||||
patch_dims = patches.shape[-1]
|
||||
patches = tf.reshape(patches, [batch_size, -1, patch_dims])
|
||||
return patches
|
||||
def get_config(self):
|
||||
|
||||
config = super().get_config().copy()
|
||||
config.update({
|
||||
'patch_size': self.patch_size,
|
||||
})
|
||||
return config
|
||||
|
|
@ -12,7 +12,7 @@ from .utils import crop_image_inside_box
|
|||
from .utils.rotate import rotate_image_different
|
||||
from .utils.resize import resize_image
|
||||
|
||||
class EynollahPlotter:
|
||||
class EynollahPlotter():
|
||||
"""
|
||||
Class collecting all the plotting and image writing methods
|
||||
"""
|
||||
|
|
@ -40,8 +40,8 @@ class EynollahPlotter:
|
|||
self.image_filename_stem = image_filename_stem
|
||||
# XXX TODO hacky these cannot be set at init time
|
||||
self.image_org = image_org
|
||||
self.scale_x : float = scale_x
|
||||
self.scale_y : float = scale_y
|
||||
self.scale_x = scale_x
|
||||
self.scale_y = scale_y
|
||||
|
||||
def save_plot_of_layout_main(self, text_regions_p, image_page):
|
||||
if self.dir_of_layout is not None:
|
||||
|
|
|
|||
|
|
@ -1,9 +1,6 @@
|
|||
from functools import cached_property
|
||||
from typing import Optional
|
||||
from ocrd_models import OcrdPage
|
||||
from ocrd import OcrdPageResultImage, Processor, OcrdPageResult
|
||||
|
||||
from eynollah.model_zoo.model_zoo import EynollahModelZoo
|
||||
from ocrd import Processor, OcrdPageResult
|
||||
|
||||
from .eynollah import Eynollah, EynollahXmlWriter
|
||||
|
||||
|
|
@ -12,25 +9,27 @@ class EynollahProcessor(Processor):
|
|||
# already employs GPU (without singleton process atm)
|
||||
max_workers = 1
|
||||
|
||||
@cached_property
|
||||
def executable(self) -> str:
|
||||
@property
|
||||
def executable(self):
|
||||
return 'ocrd-eynollah-segment'
|
||||
|
||||
def setup(self) -> None:
|
||||
assert self.parameter
|
||||
model_zoo = EynollahModelZoo(basedir=self.parameter['models'])
|
||||
if self.parameter['textline_light'] and not self.parameter['light_version']:
|
||||
raise ValueError("Error: You set parameter 'textline_light' to enable light textline detection, "
|
||||
"but parameter 'light_version' is not enabled")
|
||||
self.eynollah = Eynollah(
|
||||
model_zoo=model_zoo,
|
||||
self.resolve_resource(self.parameter['models']),
|
||||
logger=self.logger,
|
||||
allow_enhancement=self.parameter['allow_enhancement'],
|
||||
curved_line=self.parameter['curved_line'],
|
||||
right2left=self.parameter['right_to_left'],
|
||||
reading_order_machine_based=self.parameter['reading_order_machine_based'],
|
||||
ignore_page_extraction=self.parameter['ignore_page_extraction'],
|
||||
light_version=self.parameter['light_version'],
|
||||
textline_light=self.parameter['textline_light'],
|
||||
full_layout=self.parameter['full_layout'],
|
||||
allow_scaling=self.parameter['allow_scaling'],
|
||||
headers_off=self.parameter['headers_off'],
|
||||
tables=self.parameter['tables'],
|
||||
logger=self.logger
|
||||
)
|
||||
self.eynollah.plotter = None
|
||||
|
||||
|
|
@ -57,8 +56,6 @@ class EynollahProcessor(Processor):
|
|||
- If ``ignore_page_extraction``, then attempt no cropping of the page.
|
||||
- If ``curved_line``, then compute contour polygons for text lines
|
||||
instead of simple bounding boxes.
|
||||
- If ``reading_order_machine_based``, then detect reading order via
|
||||
data-driven model instead of geometrical heuristics.
|
||||
|
||||
Produce a new output file by serialising the resulting hierarchy.
|
||||
"""
|
||||
|
|
@ -88,6 +85,7 @@ class EynollahProcessor(Processor):
|
|||
dir_out=None,
|
||||
image_filename=image_filename,
|
||||
curved_line=self.eynollah.curved_line,
|
||||
textline_light=self.eynollah.textline_light,
|
||||
pcgts=pcgts)
|
||||
self.eynollah.run_single()
|
||||
return result
|
||||
|
|
|
|||
|
|
@ -2,57 +2,60 @@
|
|||
Tool to load model and binarize a given image.
|
||||
"""
|
||||
|
||||
# pyright: reportIndexIssue=false
|
||||
# pyright: reportCallIssue=false
|
||||
# pyright: reportArgumentType=false
|
||||
# pyright: reportPossiblyUnboundVariable=false
|
||||
|
||||
import sys
|
||||
from glob import glob
|
||||
import os
|
||||
import logging
|
||||
from typing import Optional
|
||||
|
||||
import numpy as np
|
||||
from PIL import Image
|
||||
import cv2
|
||||
from ocrd_utils import tf_disable_interactive_logs
|
||||
|
||||
from eynollah.model_zoo import EynollahModelZoo
|
||||
tf_disable_interactive_logs()
|
||||
import tensorflow as tf
|
||||
from tensorflow.keras.models import load_model
|
||||
from tensorflow.python.keras import backend as tensorflow_backend
|
||||
from pathlib import Path
|
||||
from .utils import is_image_filename
|
||||
|
||||
|
||||
def resize_image(img_in, input_height, input_width):
|
||||
return cv2.resize(img_in, (input_width, input_height), interpolation=cv2.INTER_NEAREST)
|
||||
|
||||
class SbbBinarizer:
|
||||
|
||||
def __init__(
|
||||
self,
|
||||
*,
|
||||
model_zoo: EynollahModelZoo,
|
||||
logger: Optional[logging.Logger] = None,
|
||||
):
|
||||
self.logger = logger if logger else logging.getLogger('eynollah.binarization')
|
||||
self.models = (model_zoo.model_path('binarization'), model_zoo.load_model('binarization'))
|
||||
self.session = self.start_new_session()
|
||||
def __init__(self, model_dir, logger=None):
|
||||
self.model_dir = model_dir
|
||||
self.log = logger if logger else logging.getLogger('SbbBinarizer')
|
||||
|
||||
self.start_new_session()
|
||||
|
||||
self.model_files = glob(self.model_dir+"/*/", recursive = True)
|
||||
|
||||
self.models = []
|
||||
for model_file in self.model_files:
|
||||
self.models.append(self.load_model(model_file))
|
||||
|
||||
def start_new_session(self):
|
||||
config = tf.compat.v1.ConfigProto()
|
||||
config.gpu_options.allow_growth = True
|
||||
|
||||
session = tf.compat.v1.Session(config=config) # tf.InteractiveSession()
|
||||
tensorflow_backend.set_session(session)
|
||||
return session
|
||||
|
||||
self.session = tf.compat.v1.Session(config=config) # tf.InteractiveSession()
|
||||
tensorflow_backend.set_session(self.session)
|
||||
|
||||
def end_session(self):
|
||||
tensorflow_backend.clear_session()
|
||||
self.session.close()
|
||||
del self.session
|
||||
|
||||
def predict(self, model, img, use_patches, n_batch_inference=5):
|
||||
def load_model(self, model_name):
|
||||
model = load_model(os.path.join(self.model_dir, model_name), compile=False)
|
||||
model_height = model.layers[len(model.layers)-1].output_shape[1]
|
||||
model_width = model.layers[len(model.layers)-1].output_shape[2]
|
||||
n_classes = model.layers[len(model.layers)-1].output_shape[3]
|
||||
return model, model_height, model_width, n_classes
|
||||
|
||||
def predict(self, model_in, img, use_patches, n_batch_inference=5):
|
||||
tensorflow_backend.set_session(self.session)
|
||||
model, model_height, model_width, n_classes = model_in
|
||||
|
||||
img_org_h = img.shape[0]
|
||||
img_org_w = img.shape[1]
|
||||
|
|
@ -311,8 +314,8 @@ class SbbBinarizer:
|
|||
prediction_true = prediction_true.astype(np.uint8)
|
||||
return prediction_true[:,:,0]
|
||||
|
||||
def run(self, image=None, image_path=None, output=None, use_patches=False, dir_in=None):
|
||||
# print(dir_in,'dir_in')
|
||||
def run(self, image=None, image_path=None, save=None, use_patches=False, dir_in=None, dir_out=None):
|
||||
print(dir_in,'dir_in')
|
||||
if not dir_in:
|
||||
if (image is not None and image_path is not None) or \
|
||||
(image is None and image_path is None):
|
||||
|
|
@ -320,38 +323,9 @@ class SbbBinarizer:
|
|||
if image_path is not None:
|
||||
image = cv2.imread(image_path)
|
||||
img_last = 0
|
||||
model_file, model = self.models
|
||||
self.logger.info('Predicting %s with model %s', image_path if image_path else '[image]', model_file)
|
||||
res = self.predict(model, image, use_patches)
|
||||
for n, (model, model_file) in enumerate(zip(self.models, self.model_files)):
|
||||
self.log.info('Predicting with model %s [%s/%s]' % (model_file, n + 1, len(self.model_files)))
|
||||
|
||||
img_fin = np.zeros((res.shape[0], res.shape[1], 3))
|
||||
res[:, :][res[:, :] == 0] = 2
|
||||
res = res - 1
|
||||
res = res * 255
|
||||
img_fin[:, :, 0] = res
|
||||
img_fin[:, :, 1] = res
|
||||
img_fin[:, :, 2] = res
|
||||
|
||||
img_fin = img_fin.astype(np.uint8)
|
||||
img_fin = (res[:, :] == 0) * 255
|
||||
img_last = img_last + img_fin
|
||||
|
||||
img_last[:, :][img_last[:, :] > 0] = 255
|
||||
img_last = (img_last[:, :] == 0) * 255
|
||||
if output:
|
||||
self.logger.info('Writing binarized image to %s', output)
|
||||
cv2.imwrite(output, img_last)
|
||||
return img_last
|
||||
else:
|
||||
ls_imgs = list(filter(is_image_filename, os.listdir(dir_in)))
|
||||
self.logger.info("Found %d image files to binarize in %s", len(ls_imgs), dir_in)
|
||||
for i, image_path in enumerate(ls_imgs):
|
||||
self.logger.info('Binarizing [%3d/%d] %s', i + 1, len(ls_imgs), image_path)
|
||||
image_stem = Path(image_path).stem
|
||||
image = cv2.imread(os.path.join(dir_in,image_path) )
|
||||
img_last = 0
|
||||
model_file, model = self.models
|
||||
self.logger.info('Predicting %s with model %s', image_path if image_path else '[image]', model_file)
|
||||
res = self.predict(model, image, use_patches)
|
||||
|
||||
img_fin = np.zeros((res.shape[0], res.shape[1], 3))
|
||||
|
|
@ -366,9 +340,38 @@ class SbbBinarizer:
|
|||
img_fin = (res[:, :] == 0) * 255
|
||||
img_last = img_last + img_fin
|
||||
|
||||
kernel = np.ones((5, 5), np.uint8)
|
||||
img_last[:, :][img_last[:, :] > 0] = 255
|
||||
img_last = (img_last[:, :] == 0) * 255
|
||||
if save:
|
||||
cv2.imwrite(save, img_last)
|
||||
return img_last
|
||||
else:
|
||||
ls_imgs = os.listdir(dir_in)
|
||||
for image_name in ls_imgs:
|
||||
image_stem = image_name.split('.')[0]
|
||||
print(image_name,'image_name')
|
||||
image = cv2.imread(os.path.join(dir_in,image_name) )
|
||||
img_last = 0
|
||||
for n, (model, model_file) in enumerate(zip(self.models, self.model_files)):
|
||||
self.log.info('Predicting with model %s [%s/%s]' % (model_file, n + 1, len(self.model_files)))
|
||||
|
||||
res = self.predict(model, image, use_patches)
|
||||
|
||||
img_fin = np.zeros((res.shape[0], res.shape[1], 3))
|
||||
res[:, :][res[:, :] == 0] = 2
|
||||
res = res - 1
|
||||
res = res * 255
|
||||
img_fin[:, :, 0] = res
|
||||
img_fin[:, :, 1] = res
|
||||
img_fin[:, :, 2] = res
|
||||
|
||||
img_fin = img_fin.astype(np.uint8)
|
||||
img_fin = (res[:, :] == 0) * 255
|
||||
img_last = img_last + img_fin
|
||||
|
||||
kernel = np.ones((5, 5), np.uint8)
|
||||
img_last[:, :][img_last[:, :] > 0] = 255
|
||||
img_last = (img_last[:, :] == 0) * 255
|
||||
|
||||
output_filename = os.path.join(output, image_stem + '.png')
|
||||
self.logger.info('Writing binarized image to %s', output_filename)
|
||||
cv2.imwrite(output_filename, img_last)
|
||||
cv2.imwrite(os.path.join(dir_out,image_stem+'.png'), img_last)
|
||||
|
|
|
|||
|
|
@ -1,24 +0,0 @@
|
|||
import click
|
||||
import tensorflow as tf
|
||||
|
||||
from .models import resnet50_unet
|
||||
|
||||
|
||||
def configuration():
|
||||
gpu_options = tf.compat.v1.GPUOptions(allow_growth=True)
|
||||
session = tf.compat.v1.Session(config=tf.compat.v1.ConfigProto(gpu_options=gpu_options))
|
||||
|
||||
@click.command()
|
||||
def build_model_load_pretrained_weights_and_save():
|
||||
n_classes = 2
|
||||
input_height = 224
|
||||
input_width = 448
|
||||
weight_decay = 1e-6
|
||||
pretraining = False
|
||||
dir_of_weights = 'model_bin_sbb_ens.h5'
|
||||
|
||||
# configuration()
|
||||
|
||||
model = resnet50_unet(n_classes, input_height, input_width, weight_decay, pretraining)
|
||||
model.load_weights(dir_of_weights)
|
||||
model.save('./name_in_another_python_version.h5')
|
||||
|
|
@ -1,30 +0,0 @@
|
|||
import os
|
||||
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
|
||||
|
||||
import click
|
||||
import sys
|
||||
|
||||
from .build_model_load_pretrained_weights_and_save import build_model_load_pretrained_weights_and_save
|
||||
from .generate_gt_for_training import main as generate_gt_cli
|
||||
from .inference import main as inference_cli
|
||||
from .train import ex
|
||||
from .extract_line_gt import linegt_cli
|
||||
from .weights_ensembling import main as ensemble_cli
|
||||
|
||||
@click.command(context_settings=dict(
|
||||
ignore_unknown_options=True,
|
||||
))
|
||||
@click.argument('SACRED_ARGS', nargs=-1, type=click.UNPROCESSED)
|
||||
def train_cli(sacred_args):
|
||||
ex.run_commandline([sys.argv[0]] + list(sacred_args))
|
||||
|
||||
@click.group('training')
|
||||
def main():
|
||||
pass
|
||||
|
||||
main.add_command(build_model_load_pretrained_weights_and_save)
|
||||
main.add_command(generate_gt_cli, 'generate-gt')
|
||||
main.add_command(inference_cli, 'inference')
|
||||
main.add_command(train_cli, 'train')
|
||||
main.add_command(linegt_cli, 'export_textline_images_and_text')
|
||||
main.add_command(ensemble_cli, 'ensembling')
|
||||
|
|
@ -1,134 +0,0 @@
|
|||
from logging import Logger, getLogger
|
||||
from typing import Optional
|
||||
from pathlib import Path
|
||||
import os
|
||||
|
||||
import click
|
||||
import cv2
|
||||
import xml.etree.ElementTree as ET
|
||||
import numpy as np
|
||||
|
||||
from ..utils import is_image_filename
|
||||
|
||||
@click.command()
|
||||
@click.option(
|
||||
"--image",
|
||||
"-i",
|
||||
help="input image filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of input images (instead of --image)",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_xmls",
|
||||
"-dx",
|
||||
help="directory of input PAGE-XML files (in addition to --dir_in; filename stems must match the image files, with '.xml' suffix).",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--out",
|
||||
"-o",
|
||||
'dir_out',
|
||||
help="directory for output PAGE-XML files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--dataset_abbrevation",
|
||||
"-ds_pref",
|
||||
'pref_of_dataset',
|
||||
help="in the case of extracting textline and text from a xml GT file user can add an abbrevation of dataset name to generated dataset",
|
||||
)
|
||||
@click.option(
|
||||
"--do_not_mask_with_textline_contour",
|
||||
"-nmtc/-mtc",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, cropped textline images will not be masked with textline contour.",
|
||||
)
|
||||
def linegt_cli(
|
||||
image,
|
||||
dir_in,
|
||||
dir_xmls,
|
||||
dir_out,
|
||||
pref_of_dataset,
|
||||
do_not_mask_with_textline_contour,
|
||||
):
|
||||
assert bool(dir_in) ^ bool(image), "Set --dir-in or --image-filename, not both"
|
||||
if dir_in:
|
||||
ls_imgs = [
|
||||
os.path.join(dir_in, image) for image in filter(is_image_filename, os.listdir(dir_in))
|
||||
]
|
||||
else:
|
||||
assert image
|
||||
ls_imgs = [image]
|
||||
|
||||
for dir_img in ls_imgs:
|
||||
file_name = Path(dir_img).stem
|
||||
dir_xml = os.path.join(dir_xmls, file_name + '.xml')
|
||||
|
||||
img = cv2.imread(dir_img)
|
||||
|
||||
total_bb_coordinates = []
|
||||
|
||||
tree1 = ET.parse(dir_xml, parser=ET.XMLParser(encoding="utf-8"))
|
||||
root1 = tree1.getroot()
|
||||
alltags = [elem.tag for elem in root1.iter()]
|
||||
|
||||
name_space = alltags[0].split('}')[0]
|
||||
name_space = name_space.split('{')[1]
|
||||
|
||||
region_tags = np.unique([x for x in alltags if x.endswith('TextRegion')])
|
||||
|
||||
cropped_lines_region_indexer = []
|
||||
|
||||
indexer_text_region = 0
|
||||
indexer_textlines = 0
|
||||
# FIXME: non recursive, use OCR-D PAGE generateDS API. Or use an existing tool for this purpose altogether
|
||||
for nn in root1.iter(region_tags):
|
||||
for child_textregion in nn:
|
||||
if child_textregion.tag.endswith("TextLine"):
|
||||
for child_textlines in child_textregion:
|
||||
if child_textlines.tag.endswith("Coords"):
|
||||
cropped_lines_region_indexer.append(indexer_text_region)
|
||||
p_h = child_textlines.attrib['points'].split(' ')
|
||||
textline_coords = np.array([[int(x.split(',')[0]), int(x.split(',')[1])] for x in p_h])
|
||||
|
||||
x, y, w, h = cv2.boundingRect(textline_coords)
|
||||
|
||||
total_bb_coordinates.append([x, y, w, h])
|
||||
|
||||
img_poly_on_img = np.copy(img)
|
||||
|
||||
mask_poly = np.zeros(img.shape)
|
||||
mask_poly = cv2.fillPoly(mask_poly, pts=[textline_coords], color=(1, 1, 1))
|
||||
|
||||
mask_poly = mask_poly[y : y + h, x : x + w, :]
|
||||
img_crop = img_poly_on_img[y : y + h, x : x + w, :]
|
||||
|
||||
if not do_not_mask_with_textline_contour:
|
||||
img_crop[mask_poly == 0] = 255
|
||||
|
||||
if img_crop.shape[0] == 0 or img_crop.shape[1] == 0:
|
||||
continue
|
||||
if child_textlines.tag.endswith("TextEquiv"):
|
||||
for cheild_text in child_textlines:
|
||||
if cheild_text.tag.endswith("Unicode"):
|
||||
textline_text = cheild_text.text
|
||||
if textline_text:
|
||||
base_name = os.path.join(
|
||||
dir_out, file_name + '_line_' + str(indexer_textlines)
|
||||
)
|
||||
if pref_of_dataset:
|
||||
base_name += '_' + pref_of_dataset
|
||||
if not do_not_mask_with_textline_contour:
|
||||
base_name += '_masked'
|
||||
|
||||
with open(base_name + '.txt', 'w') as text_file:
|
||||
text_file.write(textline_text)
|
||||
cv2.imwrite(base_name + '.png', img_crop)
|
||||
indexer_textlines += 1
|
||||
|
|
@ -1,583 +0,0 @@
|
|||
import click
|
||||
import json
|
||||
import os
|
||||
from tqdm import tqdm
|
||||
from pathlib import Path
|
||||
from PIL import Image, ImageDraw, ImageFont
|
||||
import cv2
|
||||
import numpy as np
|
||||
|
||||
from eynollah.training.gt_gen_utils import (
|
||||
filter_contours_area_of_image,
|
||||
find_format_of_given_filename_in_dir,
|
||||
find_new_features_of_contours,
|
||||
fit_text_single_line,
|
||||
get_content_of_dir,
|
||||
get_images_of_ground_truth,
|
||||
get_layout_contours_for_visualization,
|
||||
get_textline_contours_and_ocr_text,
|
||||
get_textline_contours_for_visualization,
|
||||
overlay_layout_on_image,
|
||||
read_xml,
|
||||
resize_image,
|
||||
visualize_image_from_contours,
|
||||
visualize_image_from_contours_layout
|
||||
)
|
||||
|
||||
@click.group()
|
||||
def main():
|
||||
pass
|
||||
|
||||
@main.command()
|
||||
@click.option(
|
||||
"--dir_xml",
|
||||
"-dx",
|
||||
help="directory of GT page-xml files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_images",
|
||||
"-di",
|
||||
help="directory of org images. If print space cropping or scaling is needed for labels it would be great to provide the original images to apply the same function on them. So if -ps is not set true or in config files no columns_width key is given this argumnet can be ignored. File stems in this directory should be the same as those in dir_xml.",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_out_images",
|
||||
"-doi",
|
||||
help="directory where the output org images after undergoing a process (like print space cropping or scaling) will be written.",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_out",
|
||||
"-do",
|
||||
help="directory where ground truth label images would be written",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--config",
|
||||
"-cfg",
|
||||
help="config file of prefered layout or use case.",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--type_output",
|
||||
"-to",
|
||||
help="this defines how output should be. A 2d image array or a 3d image array encoded with RGB color. Just pass 2d or 3d. The file will be saved one directory up. 2D image array is 3d but only information of one channel would be enough since all channels have the same values.",
|
||||
)
|
||||
@click.option(
|
||||
"--printspace",
|
||||
"-ps",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, generated labels and in the case of provided org images cropping will be imposed and cropped labels and images will be written in output directories.",
|
||||
)
|
||||
|
||||
def pagexml2label(dir_xml,dir_out,type_output,config, printspace, dir_images, dir_out_images):
|
||||
if config:
|
||||
with open(config) as f:
|
||||
config_params = json.load(f)
|
||||
else:
|
||||
print("passed")
|
||||
config_params = None
|
||||
gt_list = get_content_of_dir(dir_xml)
|
||||
get_images_of_ground_truth(gt_list,dir_xml,dir_out,type_output, config, config_params, printspace, dir_images, dir_out_images)
|
||||
|
||||
@main.command()
|
||||
@click.option(
|
||||
"--dir_imgs",
|
||||
"-dis",
|
||||
help="directory of images with high resolution.",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_out_images",
|
||||
"-dois",
|
||||
help="directory where degraded images will be written.",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--dir_out_labels",
|
||||
"-dols",
|
||||
help="directory where original images will be written as labels.",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--scales",
|
||||
"-scs",
|
||||
help="json dictionary where the scales are written.",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
def image_enhancement(dir_imgs, dir_out_images, dir_out_labels, scales):
|
||||
ls_imgs = os.listdir(dir_imgs)
|
||||
with open(scales) as f:
|
||||
scale_dict = json.load(f)
|
||||
ls_scales = scale_dict['scales']
|
||||
|
||||
for img in tqdm(ls_imgs):
|
||||
img_name = img.split('.')[0]
|
||||
img_type = img.split('.')[1]
|
||||
image = cv2.imread(os.path.join(dir_imgs, img))
|
||||
for i, scale in enumerate(ls_scales):
|
||||
height_sc = int(image.shape[0]*scale)
|
||||
width_sc = int(image.shape[1]*scale)
|
||||
|
||||
image_down_scaled = resize_image(image, height_sc, width_sc)
|
||||
image_back_to_org_scale = resize_image(image_down_scaled, image.shape[0], image.shape[1])
|
||||
|
||||
cv2.imwrite(os.path.join(dir_out_images, img_name+'_'+str(i)+'.'+img_type), image_back_to_org_scale)
|
||||
cv2.imwrite(os.path.join(dir_out_labels, img_name+'_'+str(i)+'.'+img_type), image)
|
||||
|
||||
|
||||
@main.command()
|
||||
@click.option(
|
||||
"--dir_xml",
|
||||
"-dx",
|
||||
help="directory of GT page-xml files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--dir_out_modal_image",
|
||||
"-domi",
|
||||
help="directory where ground truth images would be written",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--dir_out_classes",
|
||||
"-docl",
|
||||
help="directory where ground truth classes would be written",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--input_height",
|
||||
"-ih",
|
||||
help="input height",
|
||||
)
|
||||
@click.option(
|
||||
"--input_width",
|
||||
"-iw",
|
||||
help="input width",
|
||||
)
|
||||
@click.option(
|
||||
"--min_area_size",
|
||||
"-min",
|
||||
help="min area size of regions considered for reading order training.",
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--min_area_early",
|
||||
"-min_early",
|
||||
help="If you have already generated a training dataset using a specific minimum area value and now wish to create a dataset with a smaller minimum area value, you can avoid regenerating the previous dataset by providing the earlier minimum area value. This will ensure that only the missing data is generated.",
|
||||
)
|
||||
|
||||
def machine_based_reading_order(dir_xml, dir_out_modal_image, dir_out_classes, input_height, input_width, min_area_size, min_area_early):
|
||||
xml_files_ind = os.listdir(dir_xml)
|
||||
xml_files_ind = [ind_xml for ind_xml in xml_files_ind if ind_xml.endswith('.xml')]
|
||||
input_height = int(input_height)
|
||||
input_width = int(input_width)
|
||||
min_area = float(min_area_size)
|
||||
if min_area_early:
|
||||
min_area_early = float(min_area_early)
|
||||
|
||||
|
||||
indexer_start= 0#55166
|
||||
max_area = 1
|
||||
#min_area = 0.0001
|
||||
|
||||
for ind_xml in tqdm(xml_files_ind):
|
||||
indexer = 0
|
||||
#print(ind_xml)
|
||||
#print('########################')
|
||||
xml_file = os.path.join(dir_xml,ind_xml )
|
||||
f_name = ind_xml.split('.')[0]
|
||||
_, _, _, file_name, id_paragraph, id_header,co_text_paragraph,co_text_header,tot_region_ref,x_len, y_len,index_tot_regions,img_poly = read_xml(xml_file)
|
||||
|
||||
id_all_text = id_paragraph + id_header
|
||||
co_text_all = co_text_paragraph + co_text_header
|
||||
|
||||
|
||||
_, cy_main, x_min_main, x_max_main, y_min_main, y_max_main, _ = find_new_features_of_contours(co_text_header)
|
||||
|
||||
img_header_and_sep = np.zeros((y_len,x_len), dtype='uint8')
|
||||
|
||||
for j in range(len(cy_main)):
|
||||
img_header_and_sep[int(y_max_main[j]):int(y_max_main[j])+12,int(x_min_main[j]):int(x_max_main[j]) ] = 1
|
||||
|
||||
|
||||
texts_corr_order_index = [index_tot_regions[tot_region_ref.index(i)] for i in id_all_text ]
|
||||
texts_corr_order_index_int = [int(x) for x in texts_corr_order_index]
|
||||
|
||||
|
||||
co_text_all, texts_corr_order_index_int, regions_ar_less_than_early_min = filter_contours_area_of_image(img_poly, co_text_all, texts_corr_order_index_int, max_area, min_area, min_area_early)
|
||||
|
||||
|
||||
arg_array = np.array(range(len(texts_corr_order_index_int)))
|
||||
|
||||
labels_con = np.zeros((y_len,x_len,len(arg_array)),dtype='uint8')
|
||||
for i in range(len(co_text_all)):
|
||||
img_label = np.zeros((y_len,x_len,3),dtype='uint8')
|
||||
img_label=cv2.fillPoly(img_label, pts =[co_text_all[i]], color=(1,1,1))
|
||||
|
||||
img_label[:,:,0][img_poly[:,:,0]==5] = 2
|
||||
img_label[:,:,0][img_header_and_sep[:,:]==1] = 3
|
||||
|
||||
labels_con[:,:,i] = img_label[:,:,0]
|
||||
|
||||
labels_con = resize_image(labels_con, input_height, input_width)
|
||||
img_poly = resize_image(img_poly, input_height, input_width)
|
||||
|
||||
|
||||
for i in range(len(texts_corr_order_index_int)):
|
||||
for j in range(len(texts_corr_order_index_int)):
|
||||
if i!=j:
|
||||
if regions_ar_less_than_early_min:
|
||||
if regions_ar_less_than_early_min[i]==1:
|
||||
input_multi_visual_modal = np.zeros((input_height,input_width,3)).astype(np.int8)
|
||||
final_f_name = f_name+'_'+str(indexer+indexer_start)
|
||||
order_class_condition = texts_corr_order_index_int[i]-texts_corr_order_index_int[j]
|
||||
if order_class_condition<0:
|
||||
class_type = 1
|
||||
else:
|
||||
class_type = 0
|
||||
|
||||
input_multi_visual_modal[:,:,0] = labels_con[:,:,i]
|
||||
input_multi_visual_modal[:,:,1] = img_poly[:,:,0]
|
||||
input_multi_visual_modal[:,:,2] = labels_con[:,:,j]
|
||||
|
||||
np.save(os.path.join(dir_out_classes,final_f_name+'_missed.npy' ), class_type)
|
||||
|
||||
cv2.imwrite(os.path.join(dir_out_modal_image,final_f_name+'_missed.png' ), input_multi_visual_modal)
|
||||
indexer = indexer+1
|
||||
|
||||
else:
|
||||
input_multi_visual_modal = np.zeros((input_height,input_width,3)).astype(np.int8)
|
||||
final_f_name = f_name+'_'+str(indexer+indexer_start)
|
||||
order_class_condition = texts_corr_order_index_int[i]-texts_corr_order_index_int[j]
|
||||
if order_class_condition<0:
|
||||
class_type = 1
|
||||
else:
|
||||
class_type = 0
|
||||
|
||||
input_multi_visual_modal[:,:,0] = labels_con[:,:,i]
|
||||
input_multi_visual_modal[:,:,1] = img_poly[:,:,0]
|
||||
input_multi_visual_modal[:,:,2] = labels_con[:,:,j]
|
||||
|
||||
np.save(os.path.join(dir_out_classes,final_f_name+'.npy' ), class_type)
|
||||
|
||||
cv2.imwrite(os.path.join(dir_out_modal_image,final_f_name+'.png' ), input_multi_visual_modal)
|
||||
indexer = indexer+1
|
||||
|
||||
|
||||
@main.command()
|
||||
@click.option(
|
||||
"--xml_file",
|
||||
"-xml",
|
||||
help="xml filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_xml",
|
||||
"-dx",
|
||||
help="directory of GT page-xml files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--dir_out",
|
||||
"-o",
|
||||
help="directory where plots will be written",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--dir_imgs",
|
||||
"-di",
|
||||
help="directory where the overlayed plots will be written", )
|
||||
|
||||
def visualize_reading_order(xml_file, dir_xml, dir_out, dir_imgs):
|
||||
assert xml_file or dir_xml, "A single xml file -xml or a dir of xml files -dx is required not both of them"
|
||||
|
||||
if dir_xml:
|
||||
xml_files_ind = os.listdir(dir_xml)
|
||||
xml_files_ind = [ind_xml for ind_xml in xml_files_ind if ind_xml.endswith('.xml')]
|
||||
else:
|
||||
xml_files_ind = [xml_file]
|
||||
|
||||
indexer_start= 0#55166
|
||||
#min_area = 0.0001
|
||||
|
||||
for ind_xml in tqdm(xml_files_ind):
|
||||
indexer = 0
|
||||
#print(ind_xml)
|
||||
#print('########################')
|
||||
#xml_file = os.path.join(dir_xml,ind_xml )
|
||||
|
||||
if dir_xml:
|
||||
xml_file = os.path.join(dir_xml,ind_xml )
|
||||
f_name = Path(ind_xml).stem
|
||||
else:
|
||||
xml_file = os.path.join(ind_xml )
|
||||
f_name = Path(ind_xml).stem
|
||||
print(f_name, 'f_name')
|
||||
|
||||
#f_name = ind_xml.split('.')[0]
|
||||
_, _, _, file_name, id_paragraph, id_header,co_text_paragraph,co_text_header,tot_region_ref,x_len, y_len,index_tot_regions,img_poly = read_xml(xml_file)
|
||||
|
||||
id_all_text = id_paragraph + id_header
|
||||
co_text_all = co_text_paragraph + co_text_header
|
||||
|
||||
|
||||
cx_main, cy_main, x_min_main, x_max_main, y_min_main, y_max_main, _ = find_new_features_of_contours(co_text_all)
|
||||
|
||||
texts_corr_order_index = [int(index_tot_regions[tot_region_ref.index(i)]) for i in id_all_text ]
|
||||
#texts_corr_order_index_int = [int(x) for x in texts_corr_order_index]
|
||||
|
||||
|
||||
#cx_ordered = np.array(cx_main)[np.array(texts_corr_order_index)]
|
||||
#cx_ordered = cx_ordered.astype(np.int32)
|
||||
|
||||
cx_ordered = [int(val) for (_, val) in sorted(zip(texts_corr_order_index, cx_main), key=lambda x: \
|
||||
x[0], reverse=False)]
|
||||
#cx_ordered = cx_ordered.astype(np.int32)
|
||||
|
||||
cy_ordered = [int(val) for (_, val) in sorted(zip(texts_corr_order_index, cy_main), key=lambda x: \
|
||||
x[0], reverse=False)]
|
||||
#cy_ordered = cy_ordered.astype(np.int32)
|
||||
|
||||
|
||||
color = (0, 0, 255)
|
||||
thickness = 20
|
||||
if dir_imgs:
|
||||
layout = np.zeros( (y_len,x_len,3) )
|
||||
layout = cv2.fillPoly(layout, pts =co_text_all, color=(1,1,1))
|
||||
|
||||
img_file_name_with_format = find_format_of_given_filename_in_dir(dir_imgs, f_name)
|
||||
img = cv2.imread(os.path.join(dir_imgs, img_file_name_with_format))
|
||||
|
||||
overlayed = overlay_layout_on_image(layout, img, cx_ordered, cy_ordered, color, thickness)
|
||||
cv2.imwrite(os.path.join(dir_out, f_name+'.png'), overlayed)
|
||||
|
||||
else:
|
||||
img = np.zeros( (y_len,x_len,3) )
|
||||
img = cv2.fillPoly(img, pts =co_text_all, color=(255,0,0))
|
||||
for i in range(len(cx_ordered)-1):
|
||||
start_point = (int(cx_ordered[i]), int(cy_ordered[i]))
|
||||
end_point = (int(cx_ordered[i+1]), int(cy_ordered[i+1]))
|
||||
img = cv2.arrowedLine(img, start_point, end_point,
|
||||
color, thickness, tipLength = 0.03)
|
||||
|
||||
cv2.imwrite(os.path.join(dir_out, f_name+'.png'), img)
|
||||
|
||||
|
||||
@main.command()
|
||||
@click.option(
|
||||
"--xml_file",
|
||||
"-xml",
|
||||
help="xml filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_xml",
|
||||
"-dx",
|
||||
help="directory of GT page-xml files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--dir_out",
|
||||
"-o",
|
||||
help="directory where plots will be written",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--dir_imgs",
|
||||
"-di",
|
||||
help="directory of images where textline segmentation will be overlayed", )
|
||||
|
||||
def visualize_textline_segmentation(xml_file, dir_xml, dir_out, dir_imgs):
|
||||
assert xml_file or dir_xml, "A single xml file -xml or a dir of xml files -dx is required not both of them"
|
||||
if dir_xml:
|
||||
xml_files_ind = os.listdir(dir_xml)
|
||||
xml_files_ind = [ind_xml for ind_xml in xml_files_ind if ind_xml.endswith('.xml')]
|
||||
else:
|
||||
xml_files_ind = [xml_file]
|
||||
|
||||
for ind_xml in tqdm(xml_files_ind):
|
||||
indexer = 0
|
||||
#print(ind_xml)
|
||||
#print('########################')
|
||||
xml_file = os.path.join(dir_xml,ind_xml )
|
||||
f_name = Path(ind_xml).stem
|
||||
|
||||
img_file_name_with_format = find_format_of_given_filename_in_dir(dir_imgs, f_name)
|
||||
img = cv2.imread(os.path.join(dir_imgs, img_file_name_with_format))
|
||||
|
||||
co_tetxlines, y_len, x_len = get_textline_contours_for_visualization(xml_file)
|
||||
|
||||
added_image = visualize_image_from_contours(co_tetxlines, img)
|
||||
|
||||
cv2.imwrite(os.path.join(dir_out, f_name+'.png'), added_image)
|
||||
|
||||
|
||||
|
||||
@main.command()
|
||||
@click.option(
|
||||
"--xml_file",
|
||||
"-xml",
|
||||
help="xml filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_xml",
|
||||
"-dx",
|
||||
help="directory of GT page-xml files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--dir_out",
|
||||
"-o",
|
||||
help="directory where plots will be written",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--dir_imgs",
|
||||
"-di",
|
||||
help="directory of images where textline segmentation will be overlayed", )
|
||||
|
||||
def visualize_layout_segmentation(xml_file, dir_xml, dir_out, dir_imgs):
|
||||
assert xml_file or dir_xml, "A single xml file -xml or a dir of xml files -dx is required not both of them"
|
||||
if dir_xml:
|
||||
xml_files_ind = os.listdir(dir_xml)
|
||||
xml_files_ind = [ind_xml for ind_xml in xml_files_ind if ind_xml.endswith('.xml')]
|
||||
else:
|
||||
xml_files_ind = [xml_file]
|
||||
|
||||
for ind_xml in tqdm(xml_files_ind):
|
||||
indexer = 0
|
||||
#print(ind_xml)
|
||||
#print('########################')
|
||||
if dir_xml:
|
||||
xml_file = os.path.join(dir_xml,ind_xml )
|
||||
f_name = Path(ind_xml).stem
|
||||
else:
|
||||
xml_file = os.path.join(ind_xml )
|
||||
f_name = Path(ind_xml).stem
|
||||
print(f_name, 'f_name')
|
||||
|
||||
img_file_name_with_format = find_format_of_given_filename_in_dir(dir_imgs, f_name)
|
||||
img = cv2.imread(os.path.join(dir_imgs, img_file_name_with_format))
|
||||
|
||||
co_text, co_graphic, co_sep, co_img, co_table, co_map, co_noise, y_len, x_len = get_layout_contours_for_visualization(xml_file)
|
||||
|
||||
|
||||
added_image = visualize_image_from_contours_layout(co_text['paragraph'], co_text['header']+co_text['heading'], co_text['drop-capital'], co_sep, co_img, co_text['marginalia'], co_table, img)
|
||||
|
||||
cv2.imwrite(os.path.join(dir_out, f_name+'.png'), added_image)
|
||||
|
||||
|
||||
|
||||
|
||||
@main.command()
|
||||
@click.option(
|
||||
"--xml_file",
|
||||
"-xml",
|
||||
help="xml filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_xml",
|
||||
"-dx",
|
||||
help="directory of GT page-xml files",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
@click.option(
|
||||
"--dir_out",
|
||||
"-o",
|
||||
help="directory where plots will be written",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
|
||||
|
||||
def visualize_ocr_text(xml_file, dir_xml, dir_out):
|
||||
assert xml_file or dir_xml, "A single xml file -xml or a dir of xml files -dx is required not both of them"
|
||||
if dir_xml:
|
||||
xml_files_ind = os.listdir(dir_xml)
|
||||
xml_files_ind = [ind_xml for ind_xml in xml_files_ind if ind_xml.endswith('.xml')]
|
||||
else:
|
||||
xml_files_ind = [xml_file]
|
||||
|
||||
font_path = "Charis-7.000/Charis-Regular.ttf" # Make sure this file exists!
|
||||
font = ImageFont.truetype(font_path, 40)
|
||||
|
||||
for ind_xml in tqdm(xml_files_ind):
|
||||
indexer = 0
|
||||
#print(ind_xml)
|
||||
#print('########################')
|
||||
if dir_xml:
|
||||
xml_file = os.path.join(dir_xml,ind_xml )
|
||||
f_name = Path(ind_xml).stem
|
||||
else:
|
||||
xml_file = os.path.join(ind_xml )
|
||||
f_name = Path(ind_xml).stem
|
||||
print(f_name, 'f_name')
|
||||
|
||||
co_tetxlines, y_len, x_len, ocr_texts = get_textline_contours_and_ocr_text(xml_file)
|
||||
|
||||
total_bb_coordinates = []
|
||||
|
||||
image_text = Image.new("RGB", (x_len, y_len), "white")
|
||||
draw = ImageDraw.Draw(image_text)
|
||||
|
||||
|
||||
|
||||
for index, cnt in enumerate(co_tetxlines):
|
||||
x,y,w,h = cv2.boundingRect(cnt)
|
||||
#total_bb_coordinates.append([x,y,w,h])
|
||||
|
||||
#fit_text_single_line
|
||||
|
||||
#x_bb = bb_ind[0]
|
||||
#y_bb = bb_ind[1]
|
||||
#w_bb = bb_ind[2]
|
||||
#h_bb = bb_ind[3]
|
||||
if ocr_texts[index]:
|
||||
|
||||
|
||||
is_vertical = h > 2*w # Check orientation
|
||||
font = fit_text_single_line(draw, ocr_texts[index], font_path, w, int(h*0.4) )
|
||||
|
||||
if is_vertical:
|
||||
|
||||
vertical_font = fit_text_single_line(draw, ocr_texts[index], font_path, h, int(w * 0.8))
|
||||
|
||||
text_img = Image.new("RGBA", (h, w), (255, 255, 255, 0)) # Note: dimensions are swapped
|
||||
text_draw = ImageDraw.Draw(text_img)
|
||||
text_draw.text((0, 0), ocr_texts[index], font=vertical_font, fill="black")
|
||||
|
||||
# Rotate text image by 90 degrees
|
||||
rotated_text = text_img.rotate(90, expand=1)
|
||||
|
||||
# Calculate paste position (centered in bbox)
|
||||
paste_x = x + (w - rotated_text.width) // 2
|
||||
paste_y = y + (h - rotated_text.height) // 2
|
||||
|
||||
image_text.paste(rotated_text, (paste_x, paste_y), rotated_text) # Use rotated image as mask
|
||||
else:
|
||||
text_bbox = draw.textbbox((0, 0), ocr_texts[index], font=font)
|
||||
text_width = text_bbox[2] - text_bbox[0]
|
||||
text_height = text_bbox[3] - text_bbox[1]
|
||||
|
||||
text_x = x + (w - text_width) // 2 # Center horizontally
|
||||
text_y = y + (h - text_height) // 2 # Center vertically
|
||||
|
||||
# Draw the text
|
||||
draw.text((text_x, text_y), ocr_texts[index], fill="black", font=font)
|
||||
image_text.save(os.path.join(dir_out, f_name+'.png'))
|
||||
File diff suppressed because it is too large
Load diff
|
|
@ -1,725 +0,0 @@
|
|||
import sys
|
||||
import os
|
||||
from typing import Tuple
|
||||
import warnings
|
||||
import json
|
||||
|
||||
import numpy as np
|
||||
import cv2
|
||||
from numpy._typing import NDArray
|
||||
import tensorflow as tf
|
||||
from keras.models import Model, load_model
|
||||
from keras import backend as K
|
||||
import click
|
||||
from tensorflow.python.keras import backend as tensorflow_backend
|
||||
import xml.etree.ElementTree as ET
|
||||
|
||||
from .gt_gen_utils import (
|
||||
filter_contours_area_of_image,
|
||||
find_new_features_of_contours,
|
||||
read_xml,
|
||||
resize_image,
|
||||
update_list_and_return_first_with_length_bigger_than_one
|
||||
)
|
||||
from .models import (
|
||||
PatchEncoder,
|
||||
Patches
|
||||
)
|
||||
|
||||
from.utils import (scale_padd_image_for_ocr)
|
||||
from eynollah.utils.utils_ocr import (decode_batch_predictions)
|
||||
|
||||
with warnings.catch_warnings():
|
||||
warnings.simplefilter("ignore")
|
||||
|
||||
__doc__=\
|
||||
"""
|
||||
Tool to load model and predict for given image.
|
||||
"""
|
||||
|
||||
class sbb_predict:
|
||||
def __init__(self,image, dir_in, model, task, config_params_model, patches, save, save_layout, ground_truth, xml_file, cpu, out, min_area):
|
||||
self.image=image
|
||||
self.dir_in=dir_in
|
||||
self.patches=patches
|
||||
self.save=save
|
||||
self.save_layout=save_layout
|
||||
self.model_dir=model
|
||||
self.ground_truth=ground_truth
|
||||
self.task=task
|
||||
self.config_params_model=config_params_model
|
||||
self.xml_file = xml_file
|
||||
self.out = out
|
||||
self.cpu = cpu
|
||||
if min_area:
|
||||
self.min_area = float(min_area)
|
||||
else:
|
||||
self.min_area = 0
|
||||
|
||||
def resize_image(self,img_in,input_height,input_width):
|
||||
return cv2.resize( img_in, ( input_width,input_height) ,interpolation=cv2.INTER_NEAREST)
|
||||
|
||||
|
||||
def color_images(self,seg):
|
||||
ann_u=range(self.n_classes)
|
||||
if len(np.shape(seg))==3:
|
||||
seg=seg[:,:,0]
|
||||
|
||||
seg_img=np.zeros((np.shape(seg)[0],np.shape(seg)[1],3)).astype(np.uint8)
|
||||
|
||||
for c in ann_u:
|
||||
c=int(c)
|
||||
seg_img[:,:,0][seg==c]=c
|
||||
seg_img[:,:,1][seg==c]=c
|
||||
seg_img[:,:,2][seg==c]=c
|
||||
return seg_img
|
||||
|
||||
def otsu_copy_binary(self,img):
|
||||
img_r=np.zeros((img.shape[0],img.shape[1],3))
|
||||
img1=img[:,:,0]
|
||||
|
||||
#print(img.min())
|
||||
#print(img[:,:,0].min())
|
||||
#blur = cv2.GaussianBlur(img,(5,5))
|
||||
#ret3,th3 = cv2.threshold(blur,0,255,cv2.THRESH_BINARY+cv2.THRESH_OTSU)
|
||||
_, threshold1 = cv2.threshold(img1, 0, 255, cv2.THRESH_BINARY+cv2.THRESH_OTSU)
|
||||
|
||||
|
||||
|
||||
img_r[:,:,0]=threshold1
|
||||
img_r[:,:,1]=threshold1
|
||||
img_r[:,:,2]=threshold1
|
||||
#img_r=img_r/float(np.max(img_r))*255
|
||||
return img_r
|
||||
|
||||
def otsu_copy(self,img):
|
||||
img_r=np.zeros((img.shape[0],img.shape[1],3))
|
||||
#img1=img[:,:,0]
|
||||
|
||||
#print(img.min())
|
||||
#print(img[:,:,0].min())
|
||||
#blur = cv2.GaussianBlur(img,(5,5))
|
||||
#ret3,th3 = cv2.threshold(blur,0,255,cv2.THRESH_BINARY+cv2.THRESH_OTSU)
|
||||
_, threshold1 = cv2.threshold(img[:,:,0], 0, 255, cv2.THRESH_BINARY+cv2.THRESH_OTSU)
|
||||
_, threshold2 = cv2.threshold(img[:,:,1], 0, 255, cv2.THRESH_BINARY+cv2.THRESH_OTSU)
|
||||
_, threshold3 = cv2.threshold(img[:,:,2], 0, 255, cv2.THRESH_BINARY+cv2.THRESH_OTSU)
|
||||
|
||||
|
||||
|
||||
img_r[:,:,0]=threshold1
|
||||
img_r[:,:,1]=threshold2
|
||||
img_r[:,:,2]=threshold3
|
||||
###img_r=img_r/float(np.max(img_r))*255
|
||||
return img_r
|
||||
|
||||
def soft_dice_loss(self,y_true, y_pred, epsilon=1e-6):
|
||||
|
||||
axes = tuple(range(1, len(y_pred.shape)-1))
|
||||
|
||||
numerator = 2. * K.sum(y_pred * y_true, axes)
|
||||
|
||||
denominator = K.sum(K.square(y_pred) + K.square(y_true), axes)
|
||||
return 1.00 - K.mean(numerator / (denominator + epsilon)) # average over classes and batch
|
||||
|
||||
# def weighted_categorical_crossentropy(self,weights=None):
|
||||
#
|
||||
# def loss(y_true, y_pred):
|
||||
# labels_floats = tf.cast(y_true, tf.float32)
|
||||
# per_pixel_loss = tf.nn.sigmoid_cross_entropy_with_logits(labels=labels_floats,logits=y_pred)
|
||||
#
|
||||
# if weights is not None:
|
||||
# weight_mask = tf.maximum(tf.reduce_max(tf.constant(
|
||||
# np.array(weights, dtype=np.float32)[None, None, None])
|
||||
# * labels_floats, axis=-1), 1.0)
|
||||
# per_pixel_loss = per_pixel_loss * weight_mask[:, :, :, None]
|
||||
# return tf.reduce_mean(per_pixel_loss)
|
||||
# return self.loss
|
||||
|
||||
|
||||
def IoU(self,Yi,y_predi):
|
||||
## mean Intersection over Union
|
||||
## Mean IoU = TP/(FN + TP + FP)
|
||||
|
||||
IoUs = []
|
||||
Nclass = np.unique(Yi)
|
||||
for c in Nclass:
|
||||
TP = np.sum( (Yi == c)&(y_predi==c) )
|
||||
FP = np.sum( (Yi != c)&(y_predi==c) )
|
||||
FN = np.sum( (Yi == c)&(y_predi != c))
|
||||
IoU = TP/float(TP + FP + FN)
|
||||
if self.n_classes>2:
|
||||
print("class {:02.0f}: #TP={:6.0f}, #FP={:6.0f}, #FN={:5.0f}, IoU={:4.3f}".format(c,TP,FP,FN,IoU))
|
||||
IoUs.append(IoU)
|
||||
if self.n_classes>2:
|
||||
mIoU = np.mean(IoUs)
|
||||
print("_________________")
|
||||
print("Mean IoU: {:4.3f}".format(mIoU))
|
||||
return mIoU
|
||||
elif self.n_classes==2:
|
||||
mIoU = IoUs[1]
|
||||
print("_________________")
|
||||
print("IoU: {:4.3f}".format(mIoU))
|
||||
return mIoU
|
||||
|
||||
def start_new_session_and_model(self):
|
||||
if self.task == "cnn-rnn-ocr":
|
||||
if self.cpu:
|
||||
os.environ['CUDA_VISIBLE_DEVICES']='-1'
|
||||
self.model = load_model(self.model_dir)
|
||||
self.model = tf.keras.models.Model(
|
||||
self.model.get_layer(name = "image").input,
|
||||
self.model.get_layer(name = "dense2").output)
|
||||
else:
|
||||
config = tf.compat.v1.ConfigProto()
|
||||
config.gpu_options.allow_growth = True
|
||||
|
||||
session = tf.compat.v1.Session(config=config) # tf.InteractiveSession()
|
||||
tensorflow_backend.set_session(session)
|
||||
|
||||
|
||||
##if self.weights_dir!=None:
|
||||
##self.model.load_weights(self.weights_dir)
|
||||
|
||||
assert isinstance(self.model, Model)
|
||||
if self.task != 'classification' and self.task != 'reading_order':
|
||||
self.img_height=self.model.layers[len(self.model.layers)-1].output_shape[1]
|
||||
self.img_width=self.model.layers[len(self.model.layers)-1].output_shape[2]
|
||||
self.n_classes=self.model.layers[len(self.model.layers)-1].output_shape[3]
|
||||
|
||||
def visualize_model_output(self, prediction, img, task) -> Tuple[NDArray, NDArray]:
|
||||
if task == "binarization":
|
||||
prediction = prediction * -1
|
||||
prediction = prediction + 1
|
||||
added_image = prediction * 255
|
||||
layout_only = None
|
||||
else:
|
||||
unique_classes = np.unique(prediction[:,:,0])
|
||||
rgb_colors = {'0' : [255, 255, 255],
|
||||
'1' : [255, 0, 0],
|
||||
'2' : [255, 125, 0],
|
||||
'3' : [255, 0, 125],
|
||||
'4' : [125, 125, 125],
|
||||
'5' : [125, 125, 0],
|
||||
'6' : [0, 125, 255],
|
||||
'7' : [0, 125, 0],
|
||||
'8' : [125, 125, 125],
|
||||
'9' : [0, 125, 255],
|
||||
'10' : [125, 0, 125],
|
||||
'11' : [0, 255, 0],
|
||||
'12' : [0, 0, 255],
|
||||
'13' : [0, 255, 255],
|
||||
'14' : [255, 125, 125],
|
||||
'15' : [255, 0, 255]}
|
||||
|
||||
layout_only = np.zeros(prediction.shape)
|
||||
|
||||
for unq_class in unique_classes:
|
||||
rgb_class_unique = rgb_colors[str(int(unq_class))]
|
||||
layout_only[:,:,0][prediction[:,:,0]==unq_class] = rgb_class_unique[0]
|
||||
layout_only[:,:,1][prediction[:,:,0]==unq_class] = rgb_class_unique[1]
|
||||
layout_only[:,:,2][prediction[:,:,0]==unq_class] = rgb_class_unique[2]
|
||||
|
||||
|
||||
|
||||
img = self.resize_image(img, layout_only.shape[0], layout_only.shape[1])
|
||||
|
||||
layout_only = layout_only.astype(np.int32)
|
||||
img = img.astype(np.int32)
|
||||
|
||||
|
||||
|
||||
added_image = cv2.addWeighted(img,0.5,layout_only,0.1,0)
|
||||
|
||||
assert isinstance(added_image, np.ndarray)
|
||||
assert isinstance(layout_only, np.ndarray)
|
||||
return added_image, layout_only
|
||||
|
||||
def predict(self, image_dir):
|
||||
assert isinstance(self.model, Model)
|
||||
if self.task == 'classification':
|
||||
classes_names = self.config_params_model['classification_classes_name']
|
||||
img_1ch = img=cv2.imread(image_dir, 0)
|
||||
|
||||
img_1ch = img_1ch / 255.0
|
||||
img_1ch = cv2.resize(img_1ch, (self.config_params_model['input_height'], self.config_params_model['input_width']), interpolation=cv2.INTER_NEAREST)
|
||||
img_in = np.zeros((1, img_1ch.shape[0], img_1ch.shape[1], 3))
|
||||
img_in[0, :, :, 0] = img_1ch[:, :]
|
||||
img_in[0, :, :, 1] = img_1ch[:, :]
|
||||
img_in[0, :, :, 2] = img_1ch[:, :]
|
||||
|
||||
label_p_pred = self.model.predict(img_in, verbose='0')
|
||||
index_class = np.argmax(label_p_pred[0])
|
||||
|
||||
print("Predicted Class: {}".format(classes_names[str(int(index_class))]))
|
||||
elif self.task == "cnn-rnn-ocr":
|
||||
img=cv2.imread(image_dir)
|
||||
img = scale_padd_image_for_ocr(img, self.config_params_model['input_height'], self.config_params_model['input_width'])
|
||||
|
||||
img = img / 255.
|
||||
|
||||
with open(os.path.join(self.model_dir, "characters_org.txt"), 'r') as char_txt_f:
|
||||
characters = json.load(char_txt_f)
|
||||
|
||||
AUTOTUNE = tf.data.AUTOTUNE
|
||||
|
||||
# Mapping characters to integers.
|
||||
char_to_num = StringLookup(vocabulary=list(characters), mask_token=None)
|
||||
|
||||
# Mapping integers back to original characters.
|
||||
num_to_char = StringLookup(
|
||||
vocabulary=char_to_num.get_vocabulary(), mask_token=None, invert=True
|
||||
)
|
||||
preds = self.model.predict(img.reshape(1, img.shape[0], img.shape[1], img.shape[2]), verbose=0)
|
||||
pred_texts = decode_batch_predictions(preds, num_to_char)
|
||||
pred_texts = pred_texts[0].replace("[UNK]", "")
|
||||
return pred_texts
|
||||
|
||||
|
||||
elif self.task == 'reading_order':
|
||||
img_height = self.config_params_model['input_height']
|
||||
img_width = self.config_params_model['input_width']
|
||||
|
||||
tree_xml, root_xml, bb_coord_printspace, file_name, id_paragraph, id_header, co_text_paragraph, co_text_header, tot_region_ref, x_len, y_len, index_tot_regions, img_poly = read_xml(self.xml_file)
|
||||
_, cy_main, x_min_main, x_max_main, y_min_main, y_max_main, _ = find_new_features_of_contours(co_text_header)
|
||||
|
||||
img_header_and_sep = np.zeros((y_len,x_len), dtype='uint8')
|
||||
|
||||
|
||||
for j in range(len(cy_main)):
|
||||
img_header_and_sep[int(y_max_main[j]):int(y_max_main[j])+12,int(x_min_main[j]):int(x_max_main[j]) ] = 1
|
||||
|
||||
co_text_all = co_text_paragraph + co_text_header
|
||||
id_all_text = id_paragraph + id_header
|
||||
|
||||
|
||||
##texts_corr_order_index = [index_tot_regions[tot_region_ref.index(i)] for i in id_all_text ]
|
||||
##texts_corr_order_index_int = [int(x) for x in texts_corr_order_index]
|
||||
texts_corr_order_index_int = list(np.array(range(len(co_text_all))))
|
||||
|
||||
#print(texts_corr_order_index_int)
|
||||
|
||||
max_area = 1
|
||||
#print(np.shape(co_text_all[0]), len( np.shape(co_text_all[0]) ),'co_text_all')
|
||||
#co_text_all = filter_contours_area_of_image_tables(img_poly, co_text_all, _, max_area, min_area)
|
||||
#print(co_text_all,'co_text_all')
|
||||
co_text_all, texts_corr_order_index_int, _ = filter_contours_area_of_image(img_poly, co_text_all, texts_corr_order_index_int, max_area, self.min_area)
|
||||
|
||||
#print(texts_corr_order_index_int)
|
||||
|
||||
#co_text_all = [co_text_all[index] for index in texts_corr_order_index_int]
|
||||
id_all_text = [id_all_text[index] for index in texts_corr_order_index_int]
|
||||
|
||||
labels_con = np.zeros((y_len,x_len,len(co_text_all)),dtype='uint8')
|
||||
for i in range(len(co_text_all)):
|
||||
img_label = np.zeros((y_len,x_len,3),dtype='uint8')
|
||||
img_label=cv2.fillPoly(img_label, pts =[co_text_all[i]], color=(1,1,1))
|
||||
labels_con[:,:,i] = img_label[:,:,0]
|
||||
|
||||
if bb_coord_printspace:
|
||||
#bb_coord_printspace[x,y,w,h,_,_]
|
||||
x = bb_coord_printspace[0]
|
||||
y = bb_coord_printspace[1]
|
||||
w = bb_coord_printspace[2]
|
||||
h = bb_coord_printspace[3]
|
||||
labels_con = labels_con[y:y+h, x:x+w, :]
|
||||
img_poly = img_poly[y:y+h, x:x+w, :]
|
||||
img_header_and_sep = img_header_and_sep[y:y+h, x:x+w]
|
||||
|
||||
|
||||
|
||||
img3= np.copy(img_poly)
|
||||
labels_con = resize_image(labels_con, img_height, img_width)
|
||||
|
||||
img_header_and_sep = resize_image(img_header_and_sep, img_height, img_width)
|
||||
|
||||
img3= resize_image (img3, img_height, img_width)
|
||||
img3 = img3.astype(np.uint16)
|
||||
|
||||
inference_bs = 1#4
|
||||
|
||||
input_1= np.zeros( (inference_bs, img_height, img_width,3))
|
||||
|
||||
|
||||
starting_list_of_regions = [list(range(labels_con.shape[2]))]
|
||||
|
||||
index_update = 0
|
||||
index_selected = starting_list_of_regions[0]
|
||||
|
||||
scalibility_num = 0
|
||||
while index_update>=0:
|
||||
ij_list = starting_list_of_regions[index_update]
|
||||
i = ij_list[0]
|
||||
ij_list.pop(0)
|
||||
|
||||
|
||||
pr_list = []
|
||||
post_list = []
|
||||
|
||||
batch_counter = 0
|
||||
tot_counter = 1
|
||||
|
||||
tot_iteration = len(ij_list)
|
||||
full_bs_ite= tot_iteration//inference_bs
|
||||
last_bs = tot_iteration % inference_bs
|
||||
|
||||
jbatch_indexer =[]
|
||||
for j in ij_list:
|
||||
img1= np.repeat(labels_con[:,:,i][:, :, np.newaxis], 3, axis=2)
|
||||
img2 = np.repeat(labels_con[:,:,j][:, :, np.newaxis], 3, axis=2)
|
||||
|
||||
|
||||
img2[:,:,0][img3[:,:,0]==5] = 2
|
||||
img2[:,:,0][img_header_and_sep[:,:]==1] = 3
|
||||
|
||||
|
||||
|
||||
img1[:,:,0][img3[:,:,0]==5] = 2
|
||||
img1[:,:,0][img_header_and_sep[:,:]==1] = 3
|
||||
|
||||
#input_1= np.zeros( (height1, width1,3))
|
||||
|
||||
|
||||
jbatch_indexer.append(j)
|
||||
|
||||
input_1[batch_counter,:,:,0] = img1[:,:,0]/3.
|
||||
input_1[batch_counter,:,:,2] = img2[:,:,0]/3.
|
||||
input_1[batch_counter,:,:,1] = img3[:,:,0]/5.
|
||||
#input_1[batch_counter,:,:,:]= np.zeros( (batch_counter, height1, width1,3))
|
||||
batch_counter = batch_counter+1
|
||||
|
||||
#input_1[:,:,0] = img1[:,:,0]/3.
|
||||
#input_1[:,:,2] = img2[:,:,0]/3.
|
||||
#input_1[:,:,1] = img3[:,:,0]/5.
|
||||
|
||||
if batch_counter==inference_bs or ( (tot_counter//inference_bs)==full_bs_ite and tot_counter%inference_bs==last_bs):
|
||||
y_pr = self.model.predict(input_1 , verbose='0')
|
||||
scalibility_num = scalibility_num+1
|
||||
|
||||
if batch_counter==inference_bs:
|
||||
iteration_batches = inference_bs
|
||||
else:
|
||||
iteration_batches = last_bs
|
||||
for jb in range(iteration_batches):
|
||||
if y_pr[jb][0]>=0.5:
|
||||
post_list.append(jbatch_indexer[jb])
|
||||
else:
|
||||
pr_list.append(jbatch_indexer[jb])
|
||||
|
||||
batch_counter = 0
|
||||
jbatch_indexer = []
|
||||
|
||||
tot_counter = tot_counter+1
|
||||
|
||||
starting_list_of_regions, index_update = update_list_and_return_first_with_length_bigger_than_one(index_update, i, pr_list, post_list,starting_list_of_regions)
|
||||
|
||||
|
||||
index_sort = [i[0] for i in starting_list_of_regions ]
|
||||
|
||||
id_all_text = np.array(id_all_text)[index_sort]
|
||||
|
||||
alltags=[elem.tag for elem in root_xml.iter()]
|
||||
|
||||
|
||||
|
||||
link=alltags[0].split('}')[0]+'}'
|
||||
name_space = alltags[0].split('}')[0]
|
||||
name_space = name_space.split('{')[1]
|
||||
|
||||
page_element = root_xml.find(link+'Page')
|
||||
assert isinstance(page_element, ET.Element)
|
||||
|
||||
"""
|
||||
ro_subelement = ET.SubElement(page_element, 'ReadingOrder')
|
||||
#print(page_element, 'page_element')
|
||||
|
||||
#new_element = ET.Element('ReadingOrder')
|
||||
|
||||
new_element_element = ET.Element('OrderedGroup')
|
||||
new_element_element.set('id', "ro357564684568544579089")
|
||||
|
||||
for index, id_text in enumerate(id_all_text):
|
||||
new_element_2 = ET.Element('RegionRefIndexed')
|
||||
new_element_2.set('regionRef', id_all_text[index])
|
||||
new_element_2.set('index', str(index_sort[index]))
|
||||
|
||||
new_element_element.append(new_element_2)
|
||||
|
||||
ro_subelement.append(new_element_element)
|
||||
"""
|
||||
##ro_subelement = ET.SubElement(page_element, 'ReadingOrder')
|
||||
|
||||
ro_subelement = ET.Element('ReadingOrder')
|
||||
|
||||
ro_subelement2 = ET.SubElement(ro_subelement, 'OrderedGroup')
|
||||
ro_subelement2.set('id', "ro357564684568544579089")
|
||||
|
||||
for index, id_text in enumerate(id_all_text):
|
||||
new_element_2 = ET.SubElement(ro_subelement2, 'RegionRefIndexed')
|
||||
new_element_2.set('regionRef', id_all_text[index])
|
||||
new_element_2.set('index', str(index))
|
||||
|
||||
if (link+'PrintSpace' in alltags) or (link+'Border' in alltags):
|
||||
page_element.insert(1, ro_subelement)
|
||||
else:
|
||||
page_element.insert(0, ro_subelement)
|
||||
|
||||
alltags=[elem.tag for elem in root_xml.iter()]
|
||||
|
||||
ET.register_namespace("",name_space)
|
||||
tree_xml.write(os.path.join(self.out, file_name+'.xml'),xml_declaration=True,method='xml',encoding="utf8",default_namespace=None)
|
||||
#tree_xml.write('library2.xml')
|
||||
|
||||
else:
|
||||
if self.patches:
|
||||
#def textline_contours(img,input_width,input_height,n_classes,model):
|
||||
|
||||
img=cv2.imread(image_dir)
|
||||
self.img_org = np.copy(img)
|
||||
|
||||
if img.shape[0] < self.img_height:
|
||||
img = self.resize_image(img, self.img_height, img.shape[1])
|
||||
|
||||
if img.shape[1] < self.img_width:
|
||||
img = self.resize_image(img, img.shape[0], self.img_width)
|
||||
|
||||
margin = int(0.1 * self.img_width)
|
||||
width_mid = self.img_width - 2 * margin
|
||||
height_mid = self.img_height - 2 * margin
|
||||
img = img / float(255.0)
|
||||
|
||||
img_h = img.shape[0]
|
||||
img_w = img.shape[1]
|
||||
|
||||
prediction_true = np.zeros((img_h, img_w, 3))
|
||||
nxf = img_w / float(width_mid)
|
||||
nyf = img_h / float(height_mid)
|
||||
|
||||
nxf = int(nxf) + 1 if nxf > int(nxf) else int(nxf)
|
||||
nyf = int(nyf) + 1 if nyf > int(nyf) else int(nyf)
|
||||
|
||||
for i in range(nxf):
|
||||
for j in range(nyf):
|
||||
if i == 0:
|
||||
index_x_d = i * width_mid
|
||||
index_x_u = index_x_d + self.img_width
|
||||
else:
|
||||
index_x_d = i * width_mid
|
||||
index_x_u = index_x_d + self.img_width
|
||||
if j == 0:
|
||||
index_y_d = j * height_mid
|
||||
index_y_u = index_y_d + self.img_height
|
||||
else:
|
||||
index_y_d = j * height_mid
|
||||
index_y_u = index_y_d + self.img_height
|
||||
|
||||
if index_x_u > img_w:
|
||||
index_x_u = img_w
|
||||
index_x_d = img_w - self.img_width
|
||||
if index_y_u > img_h:
|
||||
index_y_u = img_h
|
||||
index_y_d = img_h - self.img_height
|
||||
|
||||
img_patch = img[index_y_d:index_y_u, index_x_d:index_x_u, :]
|
||||
label_p_pred = self.model.predict(img_patch.reshape(1, img_patch.shape[0], img_patch.shape[1], img_patch.shape[2]),
|
||||
verbose='0')
|
||||
|
||||
if self.task == 'enhancement':
|
||||
seg = label_p_pred[0, :, :, :]
|
||||
seg = seg * 255
|
||||
elif self.task == 'segmentation' or self.task == 'binarization':
|
||||
seg = np.argmax(label_p_pred, axis=3)[0]
|
||||
seg = np.repeat(seg[:, :, np.newaxis], 3, axis=2)
|
||||
else:
|
||||
raise ValueError(f"Unhandled task {self.task}")
|
||||
|
||||
|
||||
if i == 0 and j == 0:
|
||||
seg = seg[0 : seg.shape[0] - margin, 0 : seg.shape[1] - margin]
|
||||
prediction_true[index_y_d + 0 : index_y_u - margin, index_x_d + 0 : index_x_u - margin, :] = seg
|
||||
elif i == nxf - 1 and j == nyf - 1:
|
||||
seg = seg[margin : seg.shape[0] - 0, margin : seg.shape[1] - 0]
|
||||
prediction_true[index_y_d + margin : index_y_u - 0, index_x_d + margin : index_x_u - 0, :] = seg
|
||||
elif i == 0 and j == nyf - 1:
|
||||
seg = seg[margin : seg.shape[0] - 0, 0 : seg.shape[1] - margin]
|
||||
prediction_true[index_y_d + margin : index_y_u - 0, index_x_d + 0 : index_x_u - margin, :] = seg
|
||||
elif i == nxf - 1 and j == 0:
|
||||
seg = seg[0 : seg.shape[0] - margin, margin : seg.shape[1] - 0]
|
||||
prediction_true[index_y_d + 0 : index_y_u - margin, index_x_d + margin : index_x_u - 0, :] = seg
|
||||
elif i == 0 and j != 0 and j != nyf - 1:
|
||||
seg = seg[margin : seg.shape[0] - margin, 0 : seg.shape[1] - margin]
|
||||
prediction_true[index_y_d + margin : index_y_u - margin, index_x_d + 0 : index_x_u - margin, :] = seg
|
||||
elif i == nxf - 1 and j != 0 and j != nyf - 1:
|
||||
seg = seg[margin : seg.shape[0] - margin, margin : seg.shape[1] - 0]
|
||||
prediction_true[index_y_d + margin : index_y_u - margin, index_x_d + margin : index_x_u - 0, :] = seg
|
||||
elif i != 0 and i != nxf - 1 and j == 0:
|
||||
seg = seg[0 : seg.shape[0] - margin, margin : seg.shape[1] - margin]
|
||||
prediction_true[index_y_d + 0 : index_y_u - margin, index_x_d + margin : index_x_u - margin, :] = seg
|
||||
elif i != 0 and i != nxf - 1 and j == nyf - 1:
|
||||
seg = seg[margin : seg.shape[0] - 0, margin : seg.shape[1] - margin]
|
||||
prediction_true[index_y_d + margin : index_y_u - 0, index_x_d + margin : index_x_u - margin, :] = seg
|
||||
else:
|
||||
seg = seg[margin : seg.shape[0] - margin, margin : seg.shape[1] - margin]
|
||||
prediction_true[index_y_d + margin : index_y_u - margin, index_x_d + margin : index_x_u - margin, :] = seg
|
||||
prediction_true = prediction_true.astype(int)
|
||||
prediction_true = cv2.resize(prediction_true, (self.img_org.shape[1], self.img_org.shape[0]), interpolation=cv2.INTER_NEAREST)
|
||||
return prediction_true
|
||||
|
||||
else:
|
||||
|
||||
img=cv2.imread(image_dir)
|
||||
self.img_org = np.copy(img)
|
||||
|
||||
width=self.img_width
|
||||
height=self.img_height
|
||||
|
||||
img=img/255.0
|
||||
img=self.resize_image(img,self.img_height,self.img_width)
|
||||
|
||||
|
||||
label_p_pred=self.model.predict(
|
||||
img.reshape(1,img.shape[0],img.shape[1],img.shape[2]))
|
||||
|
||||
if self.task == 'enhancement':
|
||||
seg = label_p_pred[0, :, :, :]
|
||||
seg = seg * 255
|
||||
elif self.task == 'segmentation' or self.task == 'binarization':
|
||||
seg = np.argmax(label_p_pred, axis=3)[0]
|
||||
seg = np.repeat(seg[:, :, np.newaxis], 3, axis=2)
|
||||
else:
|
||||
raise ValueError(f"Unhandled task {self.task}")
|
||||
|
||||
prediction_true = seg.astype(int)
|
||||
|
||||
prediction_true = cv2.resize(prediction_true, (self.img_org.shape[1], self.img_org.shape[0]), interpolation=cv2.INTER_NEAREST)
|
||||
return prediction_true
|
||||
|
||||
|
||||
|
||||
def run(self):
|
||||
self.start_new_session_and_model()
|
||||
if self.image:
|
||||
res=self.predict(image_dir = self.image)
|
||||
|
||||
if self.task == 'classification' or self.task == 'reading_order':
|
||||
pass
|
||||
elif self.task == 'enhancement':
|
||||
if self.save:
|
||||
cv2.imwrite(self.save,res)
|
||||
elif self.task == "cnn-rnn-ocr":
|
||||
print(f"Detected text: {res}")
|
||||
else:
|
||||
img_seg_overlayed, only_layout = self.visualize_model_output(res, self.img_org, self.task)
|
||||
if self.save:
|
||||
cv2.imwrite(self.save,img_seg_overlayed)
|
||||
if self.save_layout:
|
||||
cv2.imwrite(self.save_layout, only_layout)
|
||||
|
||||
if self.ground_truth:
|
||||
gt_img=cv2.imread(self.ground_truth)
|
||||
self.IoU(gt_img[:,:,0],res[:,:,0])
|
||||
|
||||
else:
|
||||
ls_images = os.listdir(self.dir_in)
|
||||
for ind_image in ls_images:
|
||||
f_name = ind_image.split('.')[0]
|
||||
image_dir = os.path.join(self.dir_in, ind_image)
|
||||
res=self.predict(image_dir)
|
||||
|
||||
if self.task == 'classification' or self.task == 'reading_order':
|
||||
pass
|
||||
elif self.task == 'enhancement':
|
||||
self.save = os.path.join(self.out, f_name+'.png')
|
||||
cv2.imwrite(self.save,res)
|
||||
elif self.task == "cnn-rnn-ocr":
|
||||
print(f"Detected text for file name {f_name} is: {res}")
|
||||
else:
|
||||
img_seg_overlayed, only_layout = self.visualize_model_output(res, self.img_org, self.task)
|
||||
self.save = os.path.join(self.out, f_name+'_overlayed.png')
|
||||
cv2.imwrite(self.save,img_seg_overlayed)
|
||||
self.save_layout = os.path.join(self.out, f_name+'_layout.png')
|
||||
cv2.imwrite(self.save_layout, only_layout)
|
||||
|
||||
if self.ground_truth:
|
||||
gt_img=cv2.imread(self.ground_truth)
|
||||
self.IoU(gt_img[:,:,0],res[:,:,0])
|
||||
|
||||
|
||||
|
||||
@click.command()
|
||||
@click.option(
|
||||
"--image",
|
||||
"-i",
|
||||
help="image filename",
|
||||
type=click.Path(exists=True, dir_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--dir_in",
|
||||
"-di",
|
||||
help="directory of images",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--out",
|
||||
"-o",
|
||||
help="output directory where xml with detected reading order will be written.",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--patches/--no-patches",
|
||||
"-p/-nop",
|
||||
is_flag=True,
|
||||
help="if this parameter set to true, this tool will try to do inference in patches.",
|
||||
)
|
||||
@click.option(
|
||||
"--save",
|
||||
"-s",
|
||||
help="save prediction as a png file in current folder.",
|
||||
)
|
||||
@click.option(
|
||||
"--save_layout",
|
||||
"-sl",
|
||||
help="save layout prediction only as a png file in current folder.",
|
||||
)
|
||||
@click.option(
|
||||
"--model",
|
||||
"-m",
|
||||
help="directory of models",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
required=True,
|
||||
)
|
||||
@click.option(
|
||||
"--ground_truth",
|
||||
"-gt",
|
||||
help="ground truth directory if you want to see the iou of prediction.",
|
||||
)
|
||||
@click.option(
|
||||
"--xml_file",
|
||||
"-xml",
|
||||
help="xml file with layout coordinates that reading order detection will be implemented on. The result will be written in the same xml file.",
|
||||
)
|
||||
@click.option(
|
||||
"--cpu",
|
||||
"-cpu",
|
||||
help="For OCR, the default device is the GPU. If this parameter is set to true, inference will be performed on the CPU",
|
||||
is_flag=True,
|
||||
)
|
||||
@click.option(
|
||||
"--min_area",
|
||||
"-min",
|
||||
help="min area size of regions considered for reading order detection. The default value is zero and means that all text regions are considered for reading order.",
|
||||
)
|
||||
def main(image, dir_in, model, patches, save, save_layout, ground_truth, xml_file, cpu, out, min_area):
|
||||
assert image or dir_in, "Either a single image -i or a dir_in -di is required"
|
||||
with open(os.path.join(model,'config.json')) as f:
|
||||
config_params_model = json.load(f)
|
||||
task = config_params_model['task']
|
||||
if task != 'classification' and task != 'reading_order' and task != "cnn-rnn-ocr":
|
||||
if image and not save:
|
||||
print("Error: You used one of segmentation or binarization task with image input but not set -s, you need a filename to save visualized output with -s")
|
||||
sys.exit(1)
|
||||
if dir_in and not out:
|
||||
print("Error: You used one of segmentation or binarization task with dir_in but not set -out")
|
||||
sys.exit(1)
|
||||
x=sbb_predict(image, dir_in, model, task, config_params_model, patches, save, save_layout, ground_truth, xml_file, cpu, out, min_area)
|
||||
x.run()
|
||||
|
||||
|
|
@ -1,363 +0,0 @@
|
|||
from tensorflow.keras import backend as K
|
||||
import tensorflow as tf
|
||||
import numpy as np
|
||||
|
||||
|
||||
def focal_loss(gamma=2., alpha=4.):
|
||||
gamma = float(gamma)
|
||||
alpha = float(alpha)
|
||||
|
||||
def focal_loss_fixed(y_true, y_pred):
|
||||
"""Focal loss for multi-classification
|
||||
FL(p_t)=-alpha(1-p_t)^{gamma}ln(p_t)
|
||||
Notice: y_pred is probability after softmax
|
||||
gradient is d(Fl)/d(p_t) not d(Fl)/d(x) as described in paper
|
||||
d(Fl)/d(p_t) * [p_t(1-p_t)] = d(Fl)/d(x)
|
||||
Focal Loss for Dense Object Detection
|
||||
https://arxiv.org/abs/1708.02002
|
||||
|
||||
Arguments:
|
||||
y_true {tensor} -- ground truth labels, shape of [batch_size, num_cls]
|
||||
y_pred {tensor} -- model's output, shape of [batch_size, num_cls]
|
||||
|
||||
Keyword Arguments:
|
||||
gamma {float} -- (default: {2.0})
|
||||
alpha {float} -- (default: {4.0})
|
||||
|
||||
Returns:
|
||||
[tensor] -- loss.
|
||||
"""
|
||||
epsilon = 1.e-9
|
||||
y_true = tf.convert_to_tensor(y_true, tf.float32)
|
||||
y_pred = tf.convert_to_tensor(y_pred, tf.float32)
|
||||
|
||||
model_out = tf.add(y_pred, epsilon)
|
||||
ce = tf.multiply(y_true, -tf.log(model_out))
|
||||
weight = tf.multiply(y_true, tf.pow(tf.subtract(1., model_out), gamma))
|
||||
fl = tf.multiply(alpha, tf.multiply(weight, ce))
|
||||
reduced_fl = tf.reduce_max(fl, axis=1)
|
||||
return tf.reduce_mean(reduced_fl)
|
||||
|
||||
return focal_loss_fixed
|
||||
|
||||
|
||||
def weighted_categorical_crossentropy(weights=None):
|
||||
""" weighted_categorical_crossentropy
|
||||
|
||||
Args:
|
||||
* weights<ktensor|nparray|list>: crossentropy weights
|
||||
Returns:
|
||||
* weighted categorical crossentropy function
|
||||
"""
|
||||
|
||||
def loss(y_true, y_pred):
|
||||
labels_floats = tf.cast(y_true, tf.float32)
|
||||
per_pixel_loss = tf.nn.sigmoid_cross_entropy_with_logits(labels=labels_floats, logits=y_pred)
|
||||
|
||||
if weights is not None:
|
||||
weight_mask = tf.maximum(tf.reduce_max(tf.constant(
|
||||
np.array(weights, dtype=np.float32)[None, None, None])
|
||||
* labels_floats, axis=-1), 1.0)
|
||||
per_pixel_loss = per_pixel_loss * weight_mask[:, :, :, None]
|
||||
return tf.reduce_mean(per_pixel_loss)
|
||||
|
||||
return loss
|
||||
|
||||
|
||||
def image_categorical_cross_entropy(y_true, y_pred, weights=None):
|
||||
"""
|
||||
:param y_true: tensor of shape (batch_size, height, width) representing the ground truth.
|
||||
:param y_pred: tensor of shape (batch_size, height, width) representing the prediction.
|
||||
:return: The mean cross-entropy on softmaxed tensors.
|
||||
"""
|
||||
|
||||
labels_floats = tf.cast(y_true, tf.float32)
|
||||
per_pixel_loss = tf.nn.sigmoid_cross_entropy_with_logits(labels=labels_floats, logits=y_pred)
|
||||
|
||||
if weights is not None:
|
||||
weight_mask = tf.maximum(
|
||||
tf.reduce_max(tf.constant(
|
||||
np.array(weights, dtype=np.float32)[None, None, None])
|
||||
* labels_floats, axis=-1), 1.0)
|
||||
per_pixel_loss = per_pixel_loss * weight_mask[:, :, :, None]
|
||||
|
||||
return tf.reduce_mean(per_pixel_loss)
|
||||
|
||||
|
||||
def class_tversky(y_true, y_pred):
|
||||
smooth = 1.0 # 1.00
|
||||
|
||||
y_true = K.permute_dimensions(y_true, (3, 1, 2, 0))
|
||||
y_pred = K.permute_dimensions(y_pred, (3, 1, 2, 0))
|
||||
|
||||
y_true_pos = K.batch_flatten(y_true)
|
||||
y_pred_pos = K.batch_flatten(y_pred)
|
||||
true_pos = K.sum(y_true_pos * y_pred_pos, 1)
|
||||
false_neg = K.sum(y_true_pos * (1 - y_pred_pos), 1)
|
||||
false_pos = K.sum((1 - y_true_pos) * y_pred_pos, 1)
|
||||
alpha = 0.2 # 0.5
|
||||
beta = 0.8
|
||||
return (true_pos + smooth) / (true_pos + alpha * false_neg + beta * false_pos + smooth)
|
||||
|
||||
|
||||
def focal_tversky_loss(y_true, y_pred):
|
||||
pt_1 = class_tversky(y_true, y_pred)
|
||||
gamma = 1.3 # 4./3.0#1.3#4.0/3.00# 0.75
|
||||
return K.sum(K.pow((1 - pt_1), gamma))
|
||||
|
||||
|
||||
def generalized_dice_coeff2(y_true, y_pred):
|
||||
n_el = 1
|
||||
for dim in y_true.shape:
|
||||
n_el *= int(dim)
|
||||
n_cl = y_true.shape[-1]
|
||||
w = K.zeros(shape=(n_cl,))
|
||||
w = (K.sum(y_true, axis=(0, 1, 2))) / n_el
|
||||
w = 1 / (w ** 2 + 0.000001)
|
||||
numerator = y_true * y_pred
|
||||
numerator = w * K.sum(numerator, (0, 1, 2))
|
||||
numerator = K.sum(numerator)
|
||||
denominator = y_true + y_pred
|
||||
denominator = w * K.sum(denominator, (0, 1, 2))
|
||||
denominator = K.sum(denominator)
|
||||
return 2 * numerator / denominator
|
||||
|
||||
|
||||
def generalized_dice_coeff(y_true, y_pred):
|
||||
axes = tuple(range(1, len(y_pred.shape) - 1))
|
||||
Ncl = y_pred.shape[-1]
|
||||
w = K.zeros(shape=(Ncl,))
|
||||
w = K.sum(y_true, axis=axes)
|
||||
w = 1 / (w ** 2 + 0.000001)
|
||||
# Compute gen dice coef:
|
||||
numerator = y_true * y_pred
|
||||
numerator = w * K.sum(numerator, axes)
|
||||
numerator = K.sum(numerator)
|
||||
|
||||
denominator = y_true + y_pred
|
||||
denominator = w * K.sum(denominator, axes)
|
||||
denominator = K.sum(denominator)
|
||||
|
||||
gen_dice_coef = 2 * numerator / denominator
|
||||
|
||||
return gen_dice_coef
|
||||
|
||||
|
||||
def generalized_dice_loss(y_true, y_pred):
|
||||
return 1 - generalized_dice_coeff2(y_true, y_pred)
|
||||
|
||||
|
||||
# TODO: document where this is from
|
||||
def soft_dice_loss(y_true, y_pred, epsilon=1e-6):
|
||||
"""
|
||||
Soft dice loss calculation for arbitrary batch size, number of classes, and number of spatial dimensions.
|
||||
Assumes the `channels_last` format.
|
||||
|
||||
# Arguments
|
||||
y_true: b x X x Y( x Z...) x c One hot encoding of ground truth
|
||||
y_pred: b x X x Y( x Z...) x c Network output, must sum to 1 over c channel (such as after softmax)
|
||||
epsilon: Used for numerical stability to avoid divide by zero errors
|
||||
|
||||
# References
|
||||
V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation
|
||||
https://arxiv.org/abs/1606.04797
|
||||
More details on Dice loss formulation
|
||||
https://mediatum.ub.tum.de/doc/1395260/1395260.pdf (page 72)
|
||||
|
||||
Adapted from https://github.com/Lasagne/Recipes/issues/99#issuecomment-347775022
|
||||
"""
|
||||
|
||||
# skip the batch and class axis for calculating Dice score
|
||||
axes = tuple(range(1, len(y_pred.shape) - 1))
|
||||
|
||||
numerator = 2. * K.sum(y_pred * y_true, axes)
|
||||
|
||||
denominator = K.sum(K.square(y_pred) + K.square(y_true), axes)
|
||||
return 1.00 - K.mean(numerator / (denominator + epsilon)) # average over classes and batch
|
||||
|
||||
|
||||
# TODO: document where this is from
|
||||
def seg_metrics(y_true, y_pred, metric_name, metric_type='standard', drop_last=True, mean_per_class=False,
|
||||
verbose=False):
|
||||
"""
|
||||
Compute mean metrics of two segmentation masks, via Keras.
|
||||
|
||||
IoU(A,B) = |A & B| / (| A U B|)
|
||||
Dice(A,B) = 2*|A & B| / (|A| + |B|)
|
||||
|
||||
Args:
|
||||
y_true: true masks, one-hot encoded.
|
||||
y_pred: predicted masks, either softmax outputs, or one-hot encoded.
|
||||
metric_name: metric to be computed, either 'iou' or 'dice'.
|
||||
metric_type: one of 'standard' (default), 'soft', 'naive'.
|
||||
In the standard version, y_pred is one-hot encoded and the mean
|
||||
is taken only over classes that are present (in y_true or y_pred).
|
||||
The 'soft' version of the metrics are computed without one-hot
|
||||
encoding y_pred.
|
||||
The 'naive' version return mean metrics where absent classes contribute
|
||||
to the class mean as 1.0 (instead of being dropped from the mean).
|
||||
drop_last = True: boolean flag to drop last class (usually reserved
|
||||
for background class in semantic segmentation)
|
||||
mean_per_class = False: return mean along batch axis for each class.
|
||||
verbose = False: print intermediate results such as intersection, union
|
||||
(as number of pixels).
|
||||
Returns:
|
||||
IoU/Dice of y_true and y_pred, as a float, unless mean_per_class == True
|
||||
in which case it returns the per-class metric, averaged over the batch.
|
||||
|
||||
Inputs are B*W*H*N tensors, with
|
||||
B = batch size,
|
||||
W = width,
|
||||
H = height,
|
||||
N = number of classes
|
||||
"""
|
||||
|
||||
flag_soft = (metric_type == 'soft')
|
||||
flag_naive_mean = (metric_type == 'naive')
|
||||
|
||||
# always assume one or more classes
|
||||
num_classes = K.shape(y_true)[-1]
|
||||
|
||||
if not flag_soft:
|
||||
# get one-hot encoded masks from y_pred (true masks should already be one-hot)
|
||||
y_pred = K.one_hot(K.argmax(y_pred), num_classes)
|
||||
y_true = K.one_hot(K.argmax(y_true), num_classes)
|
||||
|
||||
# if already one-hot, could have skipped above command
|
||||
# keras uses float32 instead of float64, would give error down (but numpy arrays or keras.to_categorical gives float64)
|
||||
y_true = K.cast(y_true, 'float32')
|
||||
y_pred = K.cast(y_pred, 'float32')
|
||||
|
||||
# intersection and union shapes are batch_size * n_classes (values = area in pixels)
|
||||
axes = (1, 2) # W,H axes of each image
|
||||
intersection = K.sum(K.abs(y_true * y_pred), axis=axes)
|
||||
mask_sum = K.sum(K.abs(y_true), axis=axes) + K.sum(K.abs(y_pred), axis=axes)
|
||||
union = mask_sum - intersection # or, np.logical_or(y_pred, y_true) for one-hot
|
||||
|
||||
smooth = .001
|
||||
iou = (intersection + smooth) / (union + smooth)
|
||||
dice = 2 * (intersection + smooth) / (mask_sum + smooth)
|
||||
|
||||
metric = {'iou': iou, 'dice': dice}[metric_name]
|
||||
|
||||
# define mask to be 0 when no pixels are present in either y_true or y_pred, 1 otherwise
|
||||
mask = K.cast(K.not_equal(union, 0), 'float32')
|
||||
|
||||
if drop_last:
|
||||
metric = metric[:, :-1]
|
||||
mask = mask[:, :-1]
|
||||
|
||||
if verbose:
|
||||
print('intersection, union')
|
||||
print(K.eval(intersection), K.eval(union))
|
||||
print(K.eval(intersection / union))
|
||||
|
||||
# return mean metrics: remaining axes are (batch, classes)
|
||||
if flag_naive_mean:
|
||||
return K.mean(metric)
|
||||
|
||||
# take mean only over non-absent classes
|
||||
class_count = K.sum(mask, axis=0)
|
||||
non_zero = tf.greater(class_count, 0)
|
||||
non_zero_sum = tf.boolean_mask(K.sum(metric * mask, axis=0), non_zero)
|
||||
non_zero_count = tf.boolean_mask(class_count, non_zero)
|
||||
|
||||
if verbose:
|
||||
print('Counts of inputs with class present, metrics for non-absent classes')
|
||||
print(K.eval(class_count), K.eval(non_zero_sum / non_zero_count))
|
||||
|
||||
return K.mean(non_zero_sum / non_zero_count)
|
||||
|
||||
|
||||
# TODO: document where this is from
|
||||
# TODO: Why a different implementation than IoU from utils?
|
||||
def mean_iou(y_true, y_pred, **kwargs):
|
||||
"""
|
||||
Compute mean Intersection over Union of two segmentation masks, via Keras.
|
||||
|
||||
Calls metrics_k(y_true, y_pred, metric_name='iou'), see there for allowed kwargs.
|
||||
"""
|
||||
return seg_metrics(y_true, y_pred, metric_name='iou', **kwargs)
|
||||
|
||||
|
||||
def Mean_IOU(y_true, y_pred):
|
||||
nb_classes = K.int_shape(y_pred)[-1]
|
||||
iou = []
|
||||
true_pixels = K.argmax(y_true, axis=-1)
|
||||
pred_pixels = K.argmax(y_pred, axis=-1)
|
||||
void_labels = K.equal(K.sum(y_true, axis=-1), 0)
|
||||
for i in range(0, nb_classes): # exclude first label (background) and last label (void)
|
||||
true_labels = K.equal(true_pixels, i) # & ~void_labels
|
||||
pred_labels = K.equal(pred_pixels, i) # & ~void_labels
|
||||
inter = tf.to_int32(true_labels & pred_labels)
|
||||
union = tf.to_int32(true_labels | pred_labels)
|
||||
legal_batches = K.sum(tf.to_int32(true_labels), axis=1) > 0
|
||||
ious = K.sum(inter, axis=1) / K.sum(union, axis=1)
|
||||
iou.append(K.mean(tf.gather(ious, indices=tf.where(legal_batches)))) # returns average IoU of the same objects
|
||||
iou = tf.stack(iou)
|
||||
legal_labels = ~tf.debugging.is_nan(iou)
|
||||
iou = tf.gather(iou, indices=tf.where(legal_labels))
|
||||
return K.mean(iou)
|
||||
|
||||
|
||||
def iou_vahid(y_true, y_pred):
|
||||
nb_classes = tf.shape(y_true)[-1] + tf.to_int32(1)
|
||||
true_pixels = K.argmax(y_true, axis=-1)
|
||||
pred_pixels = K.argmax(y_pred, axis=-1)
|
||||
iou = []
|
||||
|
||||
for i in tf.range(nb_classes):
|
||||
tp = K.sum(tf.to_int32(K.equal(true_pixels, i) & K.equal(pred_pixels, i)))
|
||||
fp = K.sum(tf.to_int32(K.not_equal(true_pixels, i) & K.equal(pred_pixels, i)))
|
||||
fn = K.sum(tf.to_int32(K.equal(true_pixels, i) & K.not_equal(pred_pixels, i)))
|
||||
iouh = tp / (tp + fp + fn)
|
||||
iou.append(iouh)
|
||||
return K.mean(iou)
|
||||
|
||||
|
||||
# TODO: copy from utils?
|
||||
def IoU_metric(Yi, y_predi):
|
||||
# mean Intersection over Union
|
||||
# Mean IoU = TP/(FN + TP + FP)
|
||||
y_predi = np.argmax(y_predi, axis=3)
|
||||
y_testi = np.argmax(Yi, axis=3)
|
||||
IoUs = []
|
||||
Nclass = int(np.max(Yi)) + 1
|
||||
for c in range(Nclass):
|
||||
TP = np.sum((Yi == c) & (y_predi == c))
|
||||
FP = np.sum((Yi != c) & (y_predi == c))
|
||||
FN = np.sum((Yi == c) & (y_predi != c))
|
||||
IoU = TP / float(TP + FP + FN)
|
||||
IoUs.append(IoU)
|
||||
return K.cast(np.mean(IoUs), dtype='float32')
|
||||
|
||||
|
||||
def IoU_metric_keras(y_true, y_pred):
|
||||
# mean Intersection over Union
|
||||
# Mean IoU = TP/(FN + TP + FP)
|
||||
init = tf.global_variables_initializer()
|
||||
sess = tf.Session()
|
||||
sess.run(init)
|
||||
|
||||
return IoU_metric(y_true.eval(session=sess), y_pred.eval(session=sess))
|
||||
|
||||
|
||||
# TODO: unused, remove?
|
||||
def jaccard_distance_loss(y_true, y_pred, smooth=100):
|
||||
"""
|
||||
Jaccard = (|X & Y|)/ (|X|+ |Y| - |X & Y|)
|
||||
= sum(|A*B|)/(sum(|A|)+sum(|B|)-sum(|A*B|))
|
||||
|
||||
The jaccard distance loss is usefull for unbalanced datasets. This has been
|
||||
shifted so it converges on 0 and is smoothed to avoid exploding or disapearing
|
||||
gradient.
|
||||
|
||||
Ref: https://en.wikipedia.org/wiki/Jaccard_index
|
||||
|
||||
@url: https://gist.github.com/wassname/f1452b748efcbeb4cb9b1d059dce6f96
|
||||
@author: wassname
|
||||
"""
|
||||
intersection = K.sum(K.abs(y_true * y_pred), axis=-1)
|
||||
sum_ = K.sum(K.abs(y_true) + K.abs(y_pred), axis=-1)
|
||||
jac = (intersection + smooth) / (sum_ - intersection + smooth)
|
||||
return (1 - jac) * smooth
|
||||
|
|
@ -1,801 +0,0 @@
|
|||
from tensorflow import keras
|
||||
from keras.layers import (
|
||||
Activation,
|
||||
Add,
|
||||
AveragePooling2D,
|
||||
BatchNormalization,
|
||||
Conv2D,
|
||||
Dense,
|
||||
Dropout,
|
||||
Embedding,
|
||||
Flatten,
|
||||
Input,
|
||||
Lambda,
|
||||
Layer,
|
||||
LayerNormalization,
|
||||
MaxPooling2D,
|
||||
MultiHeadAttention,
|
||||
UpSampling2D,
|
||||
ZeroPadding2D,
|
||||
add,
|
||||
concatenate
|
||||
)
|
||||
from keras.models import Model
|
||||
import tensorflow as tf
|
||||
# from keras import layers, models
|
||||
from keras.regularizers import l2
|
||||
|
||||
from eynollah.patch_encoder import Patches, PatchEncoder
|
||||
|
||||
##mlp_head_units = [512, 256]#[2048, 1024]
|
||||
###projection_dim = 64
|
||||
##transformer_layers = 2#8
|
||||
##num_heads = 1#4
|
||||
resnet50_Weights_path = './pretrained_model/resnet50_weights_tf_dim_ordering_tf_kernels_notop.h5'
|
||||
IMAGE_ORDERING = 'channels_last'
|
||||
MERGE_AXIS = -1
|
||||
|
||||
|
||||
class CTCLayer(tf.keras.layers.Layer):
|
||||
def __init__(self, name=None):
|
||||
super().__init__(name=name)
|
||||
self.loss_fn = tf.keras.backend.ctc_batch_cost
|
||||
|
||||
def call(self, y_true, y_pred):
|
||||
batch_len = tf.cast(tf.shape(y_true)[0], dtype="int64")
|
||||
input_length = tf.cast(tf.shape(y_pred)[1], dtype="int64")
|
||||
label_length = tf.cast(tf.shape(y_true)[1], dtype="int64")
|
||||
|
||||
input_length = input_length * tf.ones(shape=(batch_len, 1), dtype="int64")
|
||||
label_length = label_length * tf.ones(shape=(batch_len, 1), dtype="int64")
|
||||
loss = self.loss_fn(y_true, y_pred, input_length, label_length)
|
||||
self.add_loss(loss)
|
||||
|
||||
# At test time, just return the computed predictions.
|
||||
return y_pred
|
||||
|
||||
def mlp(x, hidden_units, dropout_rate):
|
||||
for units in hidden_units:
|
||||
x = Dense(units, activation=tf.nn.gelu)(x)
|
||||
x = Dropout(dropout_rate)(x)
|
||||
return x
|
||||
|
||||
def one_side_pad(x):
|
||||
x = ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING)(x)
|
||||
if IMAGE_ORDERING == 'channels_first':
|
||||
x = Lambda(lambda x: x[:, :, :-1, :-1])(x)
|
||||
elif IMAGE_ORDERING == 'channels_last':
|
||||
x = Lambda(lambda x: x[:, :-1, :-1, :])(x)
|
||||
return x
|
||||
|
||||
|
||||
def identity_block(input_tensor, kernel_size, filters, stage, block):
|
||||
"""The identity block is the block that has no conv layer at shortcut.
|
||||
# Arguments
|
||||
input_tensor: input tensor
|
||||
kernel_size: defualt 3, the kernel size of middle conv layer at main path
|
||||
filters: list of integers, the filterss of 3 conv layer at main path
|
||||
stage: integer, current stage label, used for generating layer names
|
||||
block: 'a','b'..., current block label, used for generating layer names
|
||||
# Returns
|
||||
Output tensor for the block.
|
||||
"""
|
||||
filters1, filters2, filters3 = filters
|
||||
|
||||
if IMAGE_ORDERING == 'channels_last':
|
||||
bn_axis = 3
|
||||
else:
|
||||
bn_axis = 1
|
||||
|
||||
conv_name_base = 'res' + str(stage) + block + '_branch'
|
||||
bn_name_base = 'bn' + str(stage) + block + '_branch'
|
||||
|
||||
x = Conv2D(filters1, (1, 1), data_format=IMAGE_ORDERING, name=conv_name_base + '2a')(input_tensor)
|
||||
x = BatchNormalization(axis=bn_axis, name=bn_name_base + '2a')(x)
|
||||
x = Activation('relu')(x)
|
||||
|
||||
x = Conv2D(filters2, kernel_size, data_format=IMAGE_ORDERING,
|
||||
padding='same', name=conv_name_base + '2b')(x)
|
||||
x = BatchNormalization(axis=bn_axis, name=bn_name_base + '2b')(x)
|
||||
x = Activation('relu')(x)
|
||||
|
||||
x = Conv2D(filters3, (1, 1), data_format=IMAGE_ORDERING, name=conv_name_base + '2c')(x)
|
||||
x = BatchNormalization(axis=bn_axis, name=bn_name_base + '2c')(x)
|
||||
|
||||
x = add([x, input_tensor])
|
||||
x = Activation('relu')(x)
|
||||
return x
|
||||
|
||||
|
||||
def conv_block(input_tensor, kernel_size, filters, stage, block, strides=(2, 2)):
|
||||
"""conv_block is the block that has a conv layer at shortcut
|
||||
# Arguments
|
||||
input_tensor: input tensor
|
||||
kernel_size: defualt 3, the kernel size of middle conv layer at main path
|
||||
filters: list of integers, the filterss of 3 conv layer at main path
|
||||
stage: integer, current stage label, used for generating layer names
|
||||
block: 'a','b'..., current block label, used for generating layer names
|
||||
# Returns
|
||||
Output tensor for the block.
|
||||
Note that from stage 3, the first conv layer at main path is with strides=(2,2)
|
||||
And the shortcut should have strides=(2,2) as well
|
||||
"""
|
||||
filters1, filters2, filters3 = filters
|
||||
|
||||
if IMAGE_ORDERING == 'channels_last':
|
||||
bn_axis = 3
|
||||
else:
|
||||
bn_axis = 1
|
||||
|
||||
conv_name_base = 'res' + str(stage) + block + '_branch'
|
||||
bn_name_base = 'bn' + str(stage) + block + '_branch'
|
||||
|
||||
x = Conv2D(filters1, (1, 1), data_format=IMAGE_ORDERING, strides=strides,
|
||||
name=conv_name_base + '2a')(input_tensor)
|
||||
x = BatchNormalization(axis=bn_axis, name=bn_name_base + '2a')(x)
|
||||
x = Activation('relu')(x)
|
||||
|
||||
x = Conv2D(filters2, kernel_size, data_format=IMAGE_ORDERING, padding='same',
|
||||
name=conv_name_base + '2b')(x)
|
||||
x = BatchNormalization(axis=bn_axis, name=bn_name_base + '2b')(x)
|
||||
x = Activation('relu')(x)
|
||||
|
||||
x = Conv2D(filters3, (1, 1), data_format=IMAGE_ORDERING, name=conv_name_base + '2c')(x)
|
||||
x = BatchNormalization(axis=bn_axis, name=bn_name_base + '2c')(x)
|
||||
|
||||
shortcut = Conv2D(filters3, (1, 1), data_format=IMAGE_ORDERING, strides=strides,
|
||||
name=conv_name_base + '1')(input_tensor)
|
||||
shortcut = BatchNormalization(axis=bn_axis, name=bn_name_base + '1')(shortcut)
|
||||
|
||||
x = add([x, shortcut])
|
||||
x = Activation('relu')(x)
|
||||
return x
|
||||
|
||||
|
||||
def resnet50_unet_light(n_classes, input_height=224, input_width=224, task="segmentation", weight_decay=1e-6, pretraining=False):
|
||||
assert input_height % 32 == 0
|
||||
assert input_width % 32 == 0
|
||||
|
||||
img_input = Input(shape=(input_height, input_width, 3))
|
||||
|
||||
if IMAGE_ORDERING == 'channels_last':
|
||||
bn_axis = 3
|
||||
else:
|
||||
bn_axis = 1
|
||||
|
||||
x = ZeroPadding2D((3, 3), data_format=IMAGE_ORDERING)(img_input)
|
||||
x = Conv2D(64, (7, 7), data_format=IMAGE_ORDERING, strides=(2, 2), kernel_regularizer=l2(weight_decay),
|
||||
name='conv1')(x)
|
||||
f1 = x
|
||||
|
||||
x = BatchNormalization(axis=bn_axis, name='bn_conv1')(x)
|
||||
x = Activation('relu')(x)
|
||||
x = MaxPooling2D((3, 3), data_format=IMAGE_ORDERING, strides=(2, 2))(x)
|
||||
|
||||
x = conv_block(x, 3, [64, 64, 256], stage=2, block='a', strides=(1, 1))
|
||||
x = identity_block(x, 3, [64, 64, 256], stage=2, block='b')
|
||||
x = identity_block(x, 3, [64, 64, 256], stage=2, block='c')
|
||||
f2 = one_side_pad(x)
|
||||
|
||||
x = conv_block(x, 3, [128, 128, 512], stage=3, block='a')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='b')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='c')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='d')
|
||||
f3 = x
|
||||
|
||||
x = conv_block(x, 3, [256, 256, 1024], stage=4, block='a')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='b')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='c')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='d')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='e')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='f')
|
||||
f4 = x
|
||||
|
||||
x = conv_block(x, 3, [512, 512, 2048], stage=5, block='a')
|
||||
x = identity_block(x, 3, [512, 512, 2048], stage=5, block='b')
|
||||
x = identity_block(x, 3, [512, 512, 2048], stage=5, block='c')
|
||||
f5 = x
|
||||
|
||||
if pretraining:
|
||||
model = Model(img_input, x).load_weights(resnet50_Weights_path)
|
||||
|
||||
v512_2048 = Conv2D(512, (1, 1), padding='same', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay))(f5)
|
||||
v512_2048 = (BatchNormalization(axis=bn_axis))(v512_2048)
|
||||
v512_2048 = Activation('relu')(v512_2048)
|
||||
|
||||
v512_1024 = Conv2D(512, (1, 1), padding='same', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay))(f4)
|
||||
v512_1024 = (BatchNormalization(axis=bn_axis))(v512_1024)
|
||||
v512_1024 = Activation('relu')(v512_1024)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(v512_2048)
|
||||
o = (concatenate([o, v512_1024], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(512, (3, 3), padding='valid', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, f3], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(256, (3, 3), padding='valid', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, f2], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(128, (3, 3), padding='valid', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, f1], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(64, (3, 3), padding='valid', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, img_input], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(32, (3, 3), padding='valid', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = Conv2D(n_classes, (1, 1), padding='same', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay))(o)
|
||||
if task == "segmentation":
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = (Activation('softmax'))(o)
|
||||
else:
|
||||
o = (Activation('sigmoid'))(o)
|
||||
|
||||
model = Model(img_input, o)
|
||||
return model
|
||||
|
||||
|
||||
def resnet50_unet(n_classes, input_height=224, input_width=224, task="segmentation", weight_decay=1e-6, pretraining=False):
|
||||
assert input_height % 32 == 0
|
||||
assert input_width % 32 == 0
|
||||
|
||||
img_input = Input(shape=(input_height, input_width, 3))
|
||||
|
||||
if IMAGE_ORDERING == 'channels_last':
|
||||
bn_axis = 3
|
||||
else:
|
||||
bn_axis = 1
|
||||
|
||||
x = ZeroPadding2D((3, 3), data_format=IMAGE_ORDERING)(img_input)
|
||||
x = Conv2D(64, (7, 7), data_format=IMAGE_ORDERING, strides=(2, 2), kernel_regularizer=l2(weight_decay),
|
||||
name='conv1')(x)
|
||||
f1 = x
|
||||
|
||||
x = BatchNormalization(axis=bn_axis, name='bn_conv1')(x)
|
||||
x = Activation('relu')(x)
|
||||
x = MaxPooling2D((3, 3), data_format=IMAGE_ORDERING, strides=(2, 2))(x)
|
||||
|
||||
x = conv_block(x, 3, [64, 64, 256], stage=2, block='a', strides=(1, 1))
|
||||
x = identity_block(x, 3, [64, 64, 256], stage=2, block='b')
|
||||
x = identity_block(x, 3, [64, 64, 256], stage=2, block='c')
|
||||
f2 = one_side_pad(x)
|
||||
|
||||
x = conv_block(x, 3, [128, 128, 512], stage=3, block='a')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='b')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='c')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='d')
|
||||
f3 = x
|
||||
|
||||
x = conv_block(x, 3, [256, 256, 1024], stage=4, block='a')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='b')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='c')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='d')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='e')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='f')
|
||||
f4 = x
|
||||
|
||||
x = conv_block(x, 3, [512, 512, 2048], stage=5, block='a')
|
||||
x = identity_block(x, 3, [512, 512, 2048], stage=5, block='b')
|
||||
x = identity_block(x, 3, [512, 512, 2048], stage=5, block='c')
|
||||
f5 = x
|
||||
|
||||
if pretraining:
|
||||
Model(img_input, x).load_weights(resnet50_Weights_path)
|
||||
|
||||
v1024_2048 = Conv2D(1024, (1, 1), padding='same', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay))(
|
||||
f5)
|
||||
v1024_2048 = (BatchNormalization(axis=bn_axis))(v1024_2048)
|
||||
v1024_2048 = Activation('relu')(v1024_2048)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(v1024_2048)
|
||||
o = (concatenate([o, f4], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(512, (3, 3), padding='valid', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, f3], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(256, (3, 3), padding='valid', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, f2], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(128, (3, 3), padding='valid', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, f1], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(64, (3, 3), padding='valid', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, img_input], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(32, (3, 3), padding='valid', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = Conv2D(n_classes, (1, 1), padding='same', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay))(o)
|
||||
if task == "segmentation":
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = (Activation('softmax'))(o)
|
||||
else:
|
||||
o = (Activation('sigmoid'))(o)
|
||||
|
||||
model = Model(img_input, o)
|
||||
|
||||
return model
|
||||
|
||||
|
||||
def vit_resnet50_unet(n_classes, patch_size_x, patch_size_y, num_patches, mlp_head_units=None, transformer_layers=8, num_heads =4, projection_dim = 64, input_height=224, input_width=224, task="segmentation", weight_decay=1e-6, pretraining=False):
|
||||
if mlp_head_units is None:
|
||||
mlp_head_units = [128, 64]
|
||||
inputs = Input(shape=(input_height, input_width, 3))
|
||||
|
||||
#transformer_units = [
|
||||
#projection_dim * 2,
|
||||
#projection_dim,
|
||||
#] # Size of the transformer layers
|
||||
IMAGE_ORDERING = 'channels_last'
|
||||
bn_axis=3
|
||||
|
||||
x = ZeroPadding2D((3, 3), data_format=IMAGE_ORDERING)(inputs)
|
||||
x = Conv2D(64, (7, 7), data_format=IMAGE_ORDERING, strides=(2, 2),kernel_regularizer=l2(weight_decay), name='conv1')(x)
|
||||
f1 = x
|
||||
|
||||
x = BatchNormalization(axis=bn_axis, name='bn_conv1')(x)
|
||||
x = Activation('relu')(x)
|
||||
x = MaxPooling2D((3, 3), data_format=IMAGE_ORDERING, strides=(2, 2))(x)
|
||||
|
||||
x = conv_block(x, 3, [64, 64, 256], stage=2, block='a', strides=(1, 1))
|
||||
x = identity_block(x, 3, [64, 64, 256], stage=2, block='b')
|
||||
x = identity_block(x, 3, [64, 64, 256], stage=2, block='c')
|
||||
f2 = one_side_pad(x)
|
||||
|
||||
x = conv_block(x, 3, [128, 128, 512], stage=3, block='a')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='b')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='c')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='d')
|
||||
f3 = x
|
||||
|
||||
x = conv_block(x, 3, [256, 256, 1024], stage=4, block='a')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='b')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='c')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='d')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='e')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='f')
|
||||
f4 = x
|
||||
|
||||
x = conv_block(x, 3, [512, 512, 2048], stage=5, block='a')
|
||||
x = identity_block(x, 3, [512, 512, 2048], stage=5, block='b')
|
||||
x = identity_block(x, 3, [512, 512, 2048], stage=5, block='c')
|
||||
f5 = x
|
||||
|
||||
if pretraining:
|
||||
model = Model(inputs, x).load_weights(resnet50_Weights_path)
|
||||
|
||||
#num_patches = x.shape[1]*x.shape[2]
|
||||
|
||||
#patch_size_y = input_height / x.shape[1]
|
||||
#patch_size_x = input_width / x.shape[2]
|
||||
#patch_size = patch_size_x * patch_size_y
|
||||
patches = Patches(patch_size_x, patch_size_y)(x)
|
||||
# Encode patches.
|
||||
encoded_patches = PatchEncoder(num_patches, projection_dim)(patches)
|
||||
|
||||
for _ in range(transformer_layers):
|
||||
# Layer normalization 1.
|
||||
x1 = LayerNormalization(epsilon=1e-6)(encoded_patches)
|
||||
# Create a multi-head attention layer.
|
||||
attention_output = MultiHeadAttention(
|
||||
num_heads=num_heads, key_dim=projection_dim, dropout=0.1
|
||||
)(x1, x1)
|
||||
# Skip connection 1.
|
||||
x2 = Add()([attention_output, encoded_patches])
|
||||
# Layer normalization 2.
|
||||
x3 = LayerNormalization(epsilon=1e-6)(x2)
|
||||
# MLP.
|
||||
x3 = mlp(x3, hidden_units=mlp_head_units, dropout_rate=0.1)
|
||||
# Skip connection 2.
|
||||
encoded_patches = Add()([x3, x2])
|
||||
|
||||
assert isinstance(x, Layer)
|
||||
encoded_patches = tf.reshape(encoded_patches, [-1, x.shape[1], x.shape[2] , int( projection_dim / (patch_size_x * patch_size_y) )])
|
||||
|
||||
v1024_2048 = Conv2D( 1024 , (1, 1), padding='same', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay))(encoded_patches)
|
||||
v1024_2048 = (BatchNormalization(axis=bn_axis))(v1024_2048)
|
||||
v1024_2048 = Activation('relu')(v1024_2048)
|
||||
|
||||
o = (UpSampling2D( (2, 2), data_format=IMAGE_ORDERING))(v1024_2048)
|
||||
o = (concatenate([o, f4],axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(512, (3, 3), padding='valid', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o ,f3], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(256, (3, 3), padding='valid', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, f2], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(128, (3, 3), padding='valid', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, f1], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(64, (3, 3), padding='valid', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, inputs],axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(32, (3, 3), padding='valid', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = Conv2D(n_classes, (1, 1), padding='same', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay))(o)
|
||||
if task == "segmentation":
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = (Activation('softmax'))(o)
|
||||
else:
|
||||
o = (Activation('sigmoid'))(o)
|
||||
|
||||
model = Model(inputs=inputs, outputs=o)
|
||||
|
||||
return model
|
||||
|
||||
def vit_resnet50_unet_transformer_before_cnn(n_classes, patch_size_x, patch_size_y, num_patches, mlp_head_units=None, transformer_layers=8, num_heads =4, projection_dim = 64, input_height=224, input_width=224, task="segmentation", weight_decay=1e-6, pretraining=False):
|
||||
if mlp_head_units is None:
|
||||
mlp_head_units = [128, 64]
|
||||
inputs = Input(shape=(input_height, input_width, 3))
|
||||
|
||||
##transformer_units = [
|
||||
##projection_dim * 2,
|
||||
##projection_dim,
|
||||
##] # Size of the transformer layers
|
||||
IMAGE_ORDERING = 'channels_last'
|
||||
bn_axis=3
|
||||
|
||||
patches = Patches(patch_size_x, patch_size_y)(inputs)
|
||||
# Encode patches.
|
||||
encoded_patches = PatchEncoder(num_patches, projection_dim)(patches)
|
||||
|
||||
for _ in range(transformer_layers):
|
||||
# Layer normalization 1.
|
||||
x1 = LayerNormalization(epsilon=1e-6)(encoded_patches)
|
||||
# Create a multi-head attention layer.
|
||||
attention_output = MultiHeadAttention(
|
||||
num_heads=num_heads, key_dim=projection_dim, dropout=0.1
|
||||
)(x1, x1)
|
||||
# Skip connection 1.
|
||||
x2 = Add()([attention_output, encoded_patches])
|
||||
# Layer normalization 2.
|
||||
x3 = LayerNormalization(epsilon=1e-6)(x2)
|
||||
# MLP.
|
||||
x3 = mlp(x3, hidden_units=mlp_head_units, dropout_rate=0.1)
|
||||
# Skip connection 2.
|
||||
encoded_patches = Add()([x3, x2])
|
||||
|
||||
encoded_patches = tf.reshape(encoded_patches, [-1, input_height, input_width , int( projection_dim / (patch_size_x * patch_size_y) )])
|
||||
|
||||
encoded_patches = Conv2D(3, (1, 1), padding='same', data_format=IMAGE_ORDERING, kernel_regularizer=l2(weight_decay), name='convinput')(encoded_patches)
|
||||
|
||||
x = ZeroPadding2D((3, 3), data_format=IMAGE_ORDERING)(encoded_patches)
|
||||
x = Conv2D(64, (7, 7), data_format=IMAGE_ORDERING, strides=(2, 2),kernel_regularizer=l2(weight_decay), name='conv1')(x)
|
||||
f1 = x
|
||||
|
||||
x = BatchNormalization(axis=bn_axis, name='bn_conv1')(x)
|
||||
x = Activation('relu')(x)
|
||||
x = MaxPooling2D((3, 3), data_format=IMAGE_ORDERING, strides=(2, 2))(x)
|
||||
|
||||
x = conv_block(x, 3, [64, 64, 256], stage=2, block='a', strides=(1, 1))
|
||||
x = identity_block(x, 3, [64, 64, 256], stage=2, block='b')
|
||||
x = identity_block(x, 3, [64, 64, 256], stage=2, block='c')
|
||||
f2 = one_side_pad(x)
|
||||
|
||||
x = conv_block(x, 3, [128, 128, 512], stage=3, block='a')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='b')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='c')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='d')
|
||||
f3 = x
|
||||
|
||||
x = conv_block(x, 3, [256, 256, 1024], stage=4, block='a')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='b')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='c')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='d')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='e')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='f')
|
||||
f4 = x
|
||||
|
||||
x = conv_block(x, 3, [512, 512, 2048], stage=5, block='a')
|
||||
x = identity_block(x, 3, [512, 512, 2048], stage=5, block='b')
|
||||
x = identity_block(x, 3, [512, 512, 2048], stage=5, block='c')
|
||||
f5 = x
|
||||
|
||||
if pretraining:
|
||||
model = Model(encoded_patches, x).load_weights(resnet50_Weights_path)
|
||||
|
||||
v1024_2048 = Conv2D( 1024 , (1, 1), padding='same', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay))(x)
|
||||
v1024_2048 = (BatchNormalization(axis=bn_axis))(v1024_2048)
|
||||
v1024_2048 = Activation('relu')(v1024_2048)
|
||||
|
||||
o = (UpSampling2D( (2, 2), data_format=IMAGE_ORDERING))(v1024_2048)
|
||||
o = (concatenate([o, f4],axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(512, (3, 3), padding='valid', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o ,f3], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(256, (3, 3), padding='valid', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, f2], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(128, (3, 3), padding='valid', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, f1], axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(64, (3, 3), padding='valid', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = (UpSampling2D((2, 2), data_format=IMAGE_ORDERING))(o)
|
||||
o = (concatenate([o, inputs],axis=MERGE_AXIS))
|
||||
o = (ZeroPadding2D((1, 1), data_format=IMAGE_ORDERING))(o)
|
||||
o = (Conv2D(32, (3, 3), padding='valid', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay)))(o)
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = Activation('relu')(o)
|
||||
|
||||
o = Conv2D(n_classes, (1, 1), padding='same', data_format=IMAGE_ORDERING,kernel_regularizer=l2(weight_decay))(o)
|
||||
if task == "segmentation":
|
||||
o = (BatchNormalization(axis=bn_axis))(o)
|
||||
o = (Activation('softmax'))(o)
|
||||
else:
|
||||
o = (Activation('sigmoid'))(o)
|
||||
|
||||
model = Model(inputs=inputs, outputs=o)
|
||||
|
||||
return model
|
||||
|
||||
def resnet50_classifier(n_classes,input_height=224,input_width=224,weight_decay=1e-6,pretraining=False):
|
||||
include_top=True
|
||||
assert input_height%32 == 0
|
||||
assert input_width%32 == 0
|
||||
|
||||
|
||||
img_input = Input(shape=(input_height,input_width , 3 ))
|
||||
|
||||
if IMAGE_ORDERING == 'channels_last':
|
||||
bn_axis = 3
|
||||
else:
|
||||
bn_axis = 1
|
||||
|
||||
x = ZeroPadding2D((3, 3), data_format=IMAGE_ORDERING)(img_input)
|
||||
x = Conv2D(64, (7, 7), data_format=IMAGE_ORDERING, strides=(2, 2),kernel_regularizer=l2(weight_decay), name='conv1')(x)
|
||||
f1 = x
|
||||
|
||||
x = BatchNormalization(axis=bn_axis, name='bn_conv1')(x)
|
||||
x = Activation('relu')(x)
|
||||
x = MaxPooling2D((3, 3) , data_format=IMAGE_ORDERING , strides=(2, 2))(x)
|
||||
|
||||
|
||||
x = conv_block(x, 3, [64, 64, 256], stage=2, block='a', strides=(1, 1))
|
||||
x = identity_block(x, 3, [64, 64, 256], stage=2, block='b')
|
||||
x = identity_block(x, 3, [64, 64, 256], stage=2, block='c')
|
||||
f2 = one_side_pad(x )
|
||||
|
||||
|
||||
x = conv_block(x, 3, [128, 128, 512], stage=3, block='a')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='b')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='c')
|
||||
x = identity_block(x, 3, [128, 128, 512], stage=3, block='d')
|
||||
f3 = x
|
||||
|
||||
x = conv_block(x, 3, [256, 256, 1024], stage=4, block='a')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='b')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='c')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='d')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='e')
|
||||
x = identity_block(x, 3, [256, 256, 1024], stage=4, block='f')
|
||||
f4 = x
|
||||
|
||||
x = conv_block(x, 3, [512, 512, 2048], stage=5, block='a')
|
||||
x = identity_block(x, 3, [512, 512, 2048], stage=5, block='b')
|
||||
x = identity_block(x, 3, [512, 512, 2048], stage=5, block='c')
|
||||
f5 = x
|
||||
|
||||
if pretraining:
|
||||
Model(img_input, x).load_weights(resnet50_Weights_path)
|
||||
|
||||
x = AveragePooling2D((7, 7), name='avg_pool')(x)
|
||||
x = Flatten()(x)
|
||||
|
||||
##
|
||||
x = Dense(256, activation='relu', name='fc512')(x)
|
||||
x=Dropout(0.2)(x)
|
||||
##
|
||||
x = Dense(n_classes, activation='softmax', name='fc1000')(x)
|
||||
model = Model(img_input, x)
|
||||
|
||||
|
||||
|
||||
|
||||
return model
|
||||
|
||||
def machine_based_reading_order_model(n_classes,input_height=224,input_width=224,weight_decay=1e-6,pretraining=False):
|
||||
assert input_height%32 == 0
|
||||
assert input_width%32 == 0
|
||||
|
||||
img_input = Input(shape=(input_height,input_width , 3 ))
|
||||
|
||||
if IMAGE_ORDERING == 'channels_last':
|
||||
bn_axis = 3
|
||||
else:
|
||||
bn_axis = 1
|
||||
|
||||
x1 = ZeroPadding2D((3, 3), data_format=IMAGE_ORDERING)(img_input)
|
||||
x1 = Conv2D(64, (7, 7), data_format=IMAGE_ORDERING, strides=(2, 2),kernel_regularizer=l2(weight_decay), name='conv1')(x1)
|
||||
|
||||
x1 = BatchNormalization(axis=bn_axis, name='bn_conv1')(x1)
|
||||
x1 = Activation('relu')(x1)
|
||||
x1 = MaxPooling2D((3, 3) , data_format=IMAGE_ORDERING , strides=(2, 2))(x1)
|
||||
|
||||
x1 = conv_block(x1, 3, [64, 64, 256], stage=2, block='a', strides=(1, 1))
|
||||
x1 = identity_block(x1, 3, [64, 64, 256], stage=2, block='b')
|
||||
x1 = identity_block(x1, 3, [64, 64, 256], stage=2, block='c')
|
||||
|
||||
x1 = conv_block(x1, 3, [128, 128, 512], stage=3, block='a')
|
||||
x1 = identity_block(x1, 3, [128, 128, 512], stage=3, block='b')
|
||||
x1 = identity_block(x1, 3, [128, 128, 512], stage=3, block='c')
|
||||
x1 = identity_block(x1, 3, [128, 128, 512], stage=3, block='d')
|
||||
|
||||
x1 = conv_block(x1, 3, [256, 256, 1024], stage=4, block='a')
|
||||
x1 = identity_block(x1, 3, [256, 256, 1024], stage=4, block='b')
|
||||
x1 = identity_block(x1, 3, [256, 256, 1024], stage=4, block='c')
|
||||
x1 = identity_block(x1, 3, [256, 256, 1024], stage=4, block='d')
|
||||
x1 = identity_block(x1, 3, [256, 256, 1024], stage=4, block='e')
|
||||
x1 = identity_block(x1, 3, [256, 256, 1024], stage=4, block='f')
|
||||
|
||||
x1 = conv_block(x1, 3, [512, 512, 2048], stage=5, block='a')
|
||||
x1 = identity_block(x1, 3, [512, 512, 2048], stage=5, block='b')
|
||||
x1 = identity_block(x1, 3, [512, 512, 2048], stage=5, block='c')
|
||||
|
||||
if pretraining:
|
||||
Model(img_input , x1).load_weights(resnet50_Weights_path)
|
||||
|
||||
x1 = AveragePooling2D((7, 7), name='avg_pool1')(x1)
|
||||
flattened = Flatten()(x1)
|
||||
|
||||
o = Dense(256, activation='relu', name='fc512')(flattened)
|
||||
o=Dropout(0.2)(o)
|
||||
|
||||
o = Dense(256, activation='relu', name='fc512a')(o)
|
||||
o=Dropout(0.2)(o)
|
||||
|
||||
o = Dense(n_classes, activation='sigmoid', name='fc1000')(o)
|
||||
model = Model(img_input , o)
|
||||
|
||||
return model
|
||||
|
||||
def cnn_rnn_ocr_model(image_height=None, image_width=None, n_classes=None, max_seq=None):
|
||||
input_img = tf.keras.Input(shape=(image_height, image_width, 3), name="image")
|
||||
labels = tf.keras.layers.Input(name="label", shape=(None,))
|
||||
|
||||
x = tf.keras.layers.Conv2D(64,kernel_size=(3,3),padding="same")(input_img)
|
||||
x = tf.keras.layers.BatchNormalization(name="bn1")(x)
|
||||
x = tf.keras.layers.Activation("relu", name="relu1")(x)
|
||||
x = tf.keras.layers.Conv2D(64,kernel_size=(3,3),padding="same")(x)
|
||||
x = tf.keras.layers.BatchNormalization(name="bn2")(x)
|
||||
x = tf.keras.layers.Activation("relu", name="relu2")(x)
|
||||
x = tf.keras.layers.MaxPool2D(pool_size=(1,2),strides=(1,2))(x)
|
||||
|
||||
x = tf.keras.layers.Conv2D(128,kernel_size=(3,3),padding="same")(x)
|
||||
x = tf.keras.layers.BatchNormalization(name="bn3")(x)
|
||||
x = tf.keras.layers.Activation("relu", name="relu3")(x)
|
||||
x = tf.keras.layers.Conv2D(128,kernel_size=(3,3),padding="same")(x)
|
||||
x = tf.keras.layers.BatchNormalization(name="bn4")(x)
|
||||
x = tf.keras.layers.Activation("relu", name="relu4")(x)
|
||||
x = tf.keras.layers.MaxPool2D(pool_size=(1,2),strides=(1,2))(x)
|
||||
|
||||
x = tf.keras.layers.Conv2D(256,kernel_size=(3,3),padding="same")(x)
|
||||
x = tf.keras.layers.BatchNormalization(name="bn5")(x)
|
||||
x = tf.keras.layers.Activation("relu", name="relu5")(x)
|
||||
x = tf.keras.layers.Conv2D(256,kernel_size=(3,3),padding="same")(x)
|
||||
x = tf.keras.layers.BatchNormalization(name="bn6")(x)
|
||||
x = tf.keras.layers.Activation("relu", name="relu6")(x)
|
||||
x = tf.keras.layers.MaxPool2D(pool_size=(2,2),strides=(2,2))(x)
|
||||
|
||||
x = tf.keras.layers.Conv2D(image_width,kernel_size=(3,3),padding="same")(x)
|
||||
x = tf.keras.layers.BatchNormalization(name="bn7")(x)
|
||||
x = tf.keras.layers.Activation("relu", name="relu7")(x)
|
||||
x = tf.keras.layers.Conv2D(image_width,kernel_size=(16,1))(x)
|
||||
x = tf.keras.layers.BatchNormalization(name="bn8")(x)
|
||||
x = tf.keras.layers.Activation("relu", name="relu8")(x)
|
||||
x2d = tf.keras.layers.MaxPool2D(pool_size=(1,2),strides=(1,2))(x)
|
||||
x4d = tf.keras.layers.MaxPool2D(pool_size=(1,2),strides=(1,2))(x2d)
|
||||
|
||||
|
||||
new_shape = (x.shape[1]*x.shape[2], x.shape[3])
|
||||
new_shape2 = (x2d.shape[1]*x2d.shape[2], x2d.shape[3])
|
||||
new_shape4 = (x4d.shape[1]*x4d.shape[2], x4d.shape[3])
|
||||
|
||||
x = tf.keras.layers.Reshape(target_shape=new_shape, name="reshape")(x)
|
||||
x2d = tf.keras.layers.Reshape(target_shape=new_shape2, name="reshape2")(x2d)
|
||||
x4d = tf.keras.layers.Reshape(target_shape=new_shape4, name="reshape4")(x4d)
|
||||
|
||||
|
||||
xrnnorg = tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(image_width, return_sequences=True, dropout=0.25))(x)
|
||||
xrnn2d = tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(image_width, return_sequences=True, dropout=0.25))(x2d)
|
||||
xrnn4d = tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(image_width, return_sequences=True, dropout=0.25))(x4d)
|
||||
|
||||
xrnn2d = tf.keras.layers.Reshape(target_shape=(1, xrnn2d.shape[1], xrnn2d.shape[2]), name="reshape6")(xrnn2d)
|
||||
xrnn4d = tf.keras.layers.Reshape(target_shape=(1, xrnn4d.shape[1], xrnn4d.shape[2]), name="reshape8")(xrnn4d)
|
||||
|
||||
|
||||
xrnn2dup = tf.keras.layers.UpSampling2D(size=(1, 2), interpolation="nearest")(xrnn2d)
|
||||
xrnn4dup = tf.keras.layers.UpSampling2D(size=(1, 4), interpolation="nearest")(xrnn4d)
|
||||
|
||||
xrnn2dup = tf.keras.layers.Reshape(target_shape=(xrnn2dup.shape[2], xrnn2dup.shape[3]), name="reshape10")(xrnn2dup)
|
||||
xrnn4dup = tf.keras.layers.Reshape(target_shape=(xrnn4dup.shape[2], xrnn4dup.shape[3]), name="reshape12")(xrnn4dup)
|
||||
|
||||
addition = tf.keras.layers.Add()([xrnnorg, xrnn2dup, xrnn4dup])
|
||||
|
||||
addition_rnn = tf.keras.layers.Bidirectional(tf.keras.layers.LSTM(image_width, return_sequences=True, dropout=0.25))(addition)
|
||||
|
||||
out = tf.keras.layers.Conv1D(max_seq, 1, data_format="channels_first")(addition_rnn)
|
||||
out = tf.keras.layers.BatchNormalization(name="bn9")(out)
|
||||
out = tf.keras.layers.Activation("relu", name="relu9")(out)
|
||||
#out = tf.keras.layers.Conv1D(n_classes, 1, activation='relu', data_format="channels_last")(out)
|
||||
|
||||
out = tf.keras.layers.Dense(
|
||||
n_classes, activation="softmax", name="dense2"
|
||||
)(out)
|
||||
|
||||
# Add CTC layer for calculating CTC loss at each step.
|
||||
output = CTCLayer(name="ctc_loss")(labels, out)
|
||||
|
||||
model = tf.keras.models.Model(inputs=[input_img, labels], outputs=output, name="handwriting_recognizer")
|
||||
|
||||
return model
|
||||
|
||||
|
|
@ -1,657 +0,0 @@
|
|||
import os
|
||||
import sys
|
||||
import json
|
||||
|
||||
import click
|
||||
|
||||
from eynollah.training.metrics import (
|
||||
soft_dice_loss,
|
||||
weighted_categorical_crossentropy
|
||||
)
|
||||
from eynollah.training.models import (
|
||||
PatchEncoder,
|
||||
Patches,
|
||||
machine_based_reading_order_model,
|
||||
resnet50_classifier,
|
||||
resnet50_unet,
|
||||
vit_resnet50_unet,
|
||||
vit_resnet50_unet_transformer_before_cnn,
|
||||
cnn_rnn_ocr_model
|
||||
)
|
||||
from eynollah.training.utils import (
|
||||
data_gen,
|
||||
data_gen_ocr,
|
||||
return_multiplier_based_on_augmnentations,
|
||||
generate_arrays_from_folder_reading_order,
|
||||
generate_data_from_folder_evaluation,
|
||||
generate_data_from_folder_training,
|
||||
get_one_hot,
|
||||
provide_patches,
|
||||
return_number_of_total_training_data
|
||||
)
|
||||
|
||||
os.environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
|
||||
import tensorflow as tf
|
||||
from tensorflow.compat.v1.keras.backend import set_session
|
||||
from tensorflow.keras.optimizers import SGD, Adam
|
||||
from sacred import Experiment
|
||||
from tensorflow.keras.models import load_model
|
||||
from tqdm import tqdm
|
||||
from sklearn.metrics import f1_score
|
||||
from tensorflow.keras.callbacks import Callback
|
||||
from tensorflow.keras.layers import StringLookup
|
||||
|
||||
import numpy as np
|
||||
import cv2
|
||||
|
||||
class SaveWeightsAfterSteps(Callback):
|
||||
def __init__(self, save_interval, save_path, _config):
|
||||
super(SaveWeightsAfterSteps, self).__init__()
|
||||
self.save_interval = save_interval
|
||||
self.save_path = save_path
|
||||
self.step_count = 0
|
||||
self._config = _config
|
||||
|
||||
def on_train_batch_end(self, batch, logs=None):
|
||||
self.step_count += 1
|
||||
|
||||
if self.step_count % self.save_interval ==0:
|
||||
save_file = f"{self.save_path}/model_step_{self.step_count}"
|
||||
#os.system('mkdir '+save_file)
|
||||
|
||||
self.model.save(save_file)
|
||||
|
||||
with open(os.path.join(os.path.join(self.save_path, f"model_step_{self.step_count}"),"config.json"), "w") as fp:
|
||||
json.dump(self._config, fp) # encode dict into JSON
|
||||
print(f"saved model as steps {self.step_count} to {save_file}")
|
||||
|
||||
|
||||
|
||||
def configuration():
|
||||
config = tf.compat.v1.ConfigProto()
|
||||
config.gpu_options.allow_growth = True
|
||||
session = tf.compat.v1.Session(config=config)
|
||||
set_session(session)
|
||||
|
||||
|
||||
def get_dirs_or_files(input_data):
|
||||
image_input, labels_input = os.path.join(input_data, 'images/'), os.path.join(input_data, 'labels/')
|
||||
if os.path.isdir(input_data):
|
||||
# Check if training dir exists
|
||||
assert os.path.isdir(image_input), "{} is not a directory".format(image_input)
|
||||
assert os.path.isdir(labels_input), "{} is not a directory".format(labels_input)
|
||||
return image_input, labels_input
|
||||
|
||||
|
||||
ex = Experiment(save_git_info=False)
|
||||
|
||||
|
||||
@ex.config
|
||||
def config_params():
|
||||
n_classes = None # Number of classes. In the case of binary classification this should be 2.
|
||||
n_epochs = 1 # Number of epochs.
|
||||
input_height = 224 * 1 # Height of model's input in pixels.
|
||||
input_width = 224 * 1 # Width of model's input in pixels.
|
||||
weight_decay = 1e-6 # Weight decay of l2 regularization of model layers.
|
||||
n_batch = 1 # Number of batches at each iteration.
|
||||
max_len = None # max len for ocr output.
|
||||
learning_rate = 1e-4 # Set the learning rate.
|
||||
patches = False # Divides input image into smaller patches (input size of the model) when set to true. For the model to see the full image, like page extraction, set this to false.
|
||||
augmentation = False # To apply any kind of augmentation, this parameter must be set to true.
|
||||
flip_aug = False # If true, different types of flipping will be applied to the image. Types of flips are defined with "flip_index" in config_params.json.
|
||||
blur_aug = False # If true, different types of blurring will be applied to the image. Types of blur are defined with "blur_k" in config_params.json.
|
||||
padding_white = False # If true, white padding will be applied to the image.
|
||||
padding_black = False # If true, black padding will be applied to the image.
|
||||
scaling = False # If true, scaling will be applied to the image. The amount of scaling is defined with "scales" in config_params.json.
|
||||
shifting = False
|
||||
degrading = False # If true, degrading will be applied to the image. The amount of degrading is defined with "degrade_scales" in config_params.json.
|
||||
brightening = False # If true, brightening will be applied to the image. The amount of brightening is defined with "brightness" in config_params.json.
|
||||
binarization = False # If true, Otsu thresholding will be applied to augment the input with binarized images.
|
||||
image_inversion = False # If true, and if the binarized images are avilable the image inevrsion will be applied.
|
||||
white_noise_strap = False # If true, white noise will be applied on some straps on the textline image.
|
||||
textline_skewing = False # If true, textline images will be skewed for augmentation.
|
||||
textline_skewing_bin = False # If true, textline image skewing augmentation for binarized images will be applied if already are available.
|
||||
textline_left_in_depth = False # If true, left side of textline image will be displayed in depth.
|
||||
textline_left_in_depth_bin = False # If true, left side of textline binarized image (if available) will be displayed in depth.
|
||||
textline_right_in_depth = False # If true, right side of textline image will be displayed in depth.
|
||||
textline_right_in_depth_bin = False # If true, right side of textline binarized image (if available) will be displayed in depth.
|
||||
textline_up_in_depth = False # If true, upper side of textline image will be displayed in depth.
|
||||
textline_up_in_depth_bin = False # If true, upper side of textline binarized image (if available) will be displayed in depth.
|
||||
textline_down_in_depth = False # If true, lower side of textline image will be displayed in depth.
|
||||
textline_down_in_depth_bin = False # If true, lower side of textline binarized image (if available) will be displayed in depth.
|
||||
pepper_bin_aug = False # If true, pepper noise will be added to textline binarized image (if available).
|
||||
pepper_aug = False # If true, pepper noise will be added to textline image.
|
||||
adding_rgb_background = False
|
||||
adding_rgb_foreground = False
|
||||
add_red_textlines = False
|
||||
channels_shuffling = False
|
||||
dir_train = None # Directory of training dataset with subdirectories having the names "images" and "labels".
|
||||
dir_eval = None # Directory of validation dataset with subdirectories having the names "images" and "labels".
|
||||
dir_output = None # Directory where the output model will be saved.
|
||||
pretraining = False # Set to true to load pretrained weights of ResNet50 encoder.
|
||||
scaling_bluring = False # If true, a combination of scaling and blurring will be applied to the image.
|
||||
scaling_binarization = False # If true, a combination of scaling and binarization will be applied to the image.
|
||||
bin_deg = False # If true, a combination of degrading and binarization will be applied to the image.
|
||||
rotation = False # If true, a 90 degree rotation will be implemeneted.
|
||||
color_padding_rotation = False # If true, rotation and padding will be implemeneted.
|
||||
rotation_not_90 = False # If true rotation based on provided angles with thetha will be implemeneted.
|
||||
scaling_brightness = False # If true, a combination of scaling and brightening will be applied to the image.
|
||||
scaling_flip = False # If true, a combination of scaling and flipping will be applied to the image.
|
||||
thetha = None # Rotate image by these angles for augmentation.
|
||||
thetha_padd = None # List of angles used for rotation alongside padding
|
||||
shuffle_indexes = None # List of shuffling indexes like [[0,2,1], [1,2,0], [1,0,2]]
|
||||
pepper_indexes = None # List of pepper noise indexes like [0.01, 0.005]
|
||||
white_padds = None # List of padding size in the case of white padding
|
||||
skewing_amplitudes = None # List of skewing augmentation amplitudes like [5, 8]
|
||||
blur_k = None # Blur image for augmentation.
|
||||
scales = None # Scale patches for augmentation.
|
||||
padd_colors = None # padding colors. A list elements can be only white and black. like ["white", "black"] or only one of them ["white"]
|
||||
degrade_scales = None # Degrade image for augmentation.
|
||||
brightness = None # Brighten image for augmentation.
|
||||
flip_index = None # Flip image for augmentation.
|
||||
continue_training = False # Set to true if you would like to continue training an already trained a model.
|
||||
transformer_patchsize_x = None # Patch size of vision transformer patches in x direction.
|
||||
transformer_patchsize_y = None # Patch size of vision transformer patches in y direction.
|
||||
transformer_num_patches_xy = None # Number of patches for vision transformer in x and y direction respectively.
|
||||
transformer_projection_dim = 64 # Transformer projection dimension. Default value is 64.
|
||||
transformer_mlp_head_units = [128, 64] # Transformer Multilayer Perceptron (MLP) head units. Default value is [128, 64]
|
||||
transformer_layers = 8 # transformer layers. Default value is 8.
|
||||
transformer_num_heads = 4 # Transformer number of heads. Default value is 4.
|
||||
transformer_cnn_first = True # We have two types of vision transformers. In one type, a CNN is applied first, followed by a transformer. In the other type, this order is reversed. If transformer_cnn_first is true, it means the CNN will be applied before the transformer. Default value is true.
|
||||
index_start = 0 # Index of model to continue training from. E.g. if you trained for 3 epochs and last index is 2, to continue from model_1.h5, set "index_start" to 3 to start naming model with index 3.
|
||||
dir_of_start_model = '' # Directory containing pretrained encoder to continue training the model.
|
||||
is_loss_soft_dice = False # Use soft dice as loss function. When set to true, "weighted_loss" must be false.
|
||||
weighted_loss = False # Use weighted categorical cross entropy as loss fucntion. When set to true, "is_loss_soft_dice" must be false.
|
||||
data_is_provided = False # Only set this to true when you have already provided the input data and the train and eval data are in "dir_output".
|
||||
task = "segmentation" # This parameter defines task of model which can be segmentation, enhancement or classification.
|
||||
f1_threshold_classification = None # This threshold is used to consider models with an evaluation f1 scores bigger than it. The selected model weights undergo a weights ensembling. And avreage ensembled model will be written to output.
|
||||
classification_classes_name = None # Dictionary of classification classes names.
|
||||
backbone_type = None # As backbone we have 2 types of backbones. A vision transformer alongside a CNN and we call it "transformer" and only CNN called "nontransformer"
|
||||
save_interval = None
|
||||
dir_img_bin = None
|
||||
number_of_backgrounds_per_image = 1
|
||||
dir_rgb_backgrounds = None
|
||||
dir_rgb_foregrounds = None
|
||||
characters_txt_file = None # Directory of characters text file needed for cnn_rnn_ocr model training. The file ends with .txt
|
||||
|
||||
@ex.automain
|
||||
def run(
|
||||
_config,
|
||||
n_classes,
|
||||
n_epochs,
|
||||
input_height,
|
||||
input_width,
|
||||
weight_decay,
|
||||
weighted_loss,
|
||||
index_start,
|
||||
dir_of_start_model,
|
||||
is_loss_soft_dice,
|
||||
n_batch,
|
||||
patches,
|
||||
augmentation,
|
||||
flip_aug,
|
||||
blur_aug,
|
||||
padding_white,
|
||||
padding_black,
|
||||
scaling,
|
||||
shifting,
|
||||
degrading,
|
||||
channels_shuffling,
|
||||
brightening,
|
||||
binarization,
|
||||
adding_rgb_background,
|
||||
adding_rgb_foreground,
|
||||
add_red_textlines,
|
||||
blur_k,
|
||||
scales,
|
||||
degrade_scales,
|
||||
shuffle_indexes,
|
||||
brightness,
|
||||
dir_train,
|
||||
data_is_provided,
|
||||
scaling_bluring,
|
||||
scaling_brightness,
|
||||
scaling_binarization,
|
||||
rotation,
|
||||
rotation_not_90,
|
||||
thetha,
|
||||
thetha_padd,
|
||||
scaling_flip,
|
||||
continue_training,
|
||||
transformer_projection_dim,
|
||||
transformer_mlp_head_units,
|
||||
transformer_layers,
|
||||
transformer_num_heads,
|
||||
transformer_cnn_first,
|
||||
transformer_patchsize_x,
|
||||
transformer_patchsize_y,
|
||||
transformer_num_patches_xy,
|
||||
backbone_type,
|
||||
save_interval,
|
||||
flip_index,
|
||||
dir_eval,
|
||||
dir_output,
|
||||
pretraining,
|
||||
learning_rate,
|
||||
task,
|
||||
f1_threshold_classification,
|
||||
classification_classes_name,
|
||||
dir_img_bin,
|
||||
number_of_backgrounds_per_image,
|
||||
dir_rgb_backgrounds,
|
||||
dir_rgb_foregrounds,
|
||||
characters_txt_file,
|
||||
color_padding_rotation,
|
||||
bin_deg,
|
||||
image_inversion,
|
||||
white_noise_strap,
|
||||
textline_skewing,
|
||||
textline_skewing_bin,
|
||||
textline_left_in_depth,
|
||||
textline_left_in_depth_bin,
|
||||
textline_right_in_depth,
|
||||
textline_right_in_depth_bin,
|
||||
textline_up_in_depth,
|
||||
textline_up_in_depth_bin,
|
||||
textline_down_in_depth,
|
||||
textline_down_in_depth_bin,
|
||||
pepper_bin_aug,
|
||||
pepper_aug,
|
||||
padd_colors,
|
||||
pepper_indexes,
|
||||
white_padds,
|
||||
skewing_amplitudes,
|
||||
max_len,
|
||||
):
|
||||
|
||||
if dir_rgb_backgrounds:
|
||||
list_all_possible_background_images = os.listdir(dir_rgb_backgrounds)
|
||||
else:
|
||||
list_all_possible_background_images = None
|
||||
|
||||
if dir_rgb_foregrounds:
|
||||
list_all_possible_foreground_rgbs = os.listdir(dir_rgb_foregrounds)
|
||||
else:
|
||||
list_all_possible_foreground_rgbs = None
|
||||
|
||||
dir_seg = None
|
||||
weights = None
|
||||
model = None
|
||||
|
||||
if task == "segmentation" or task == "enhancement" or task == "binarization":
|
||||
if data_is_provided:
|
||||
dir_train_flowing = os.path.join(dir_output, 'train')
|
||||
dir_eval_flowing = os.path.join(dir_output, 'eval')
|
||||
|
||||
|
||||
dir_flow_train_imgs = os.path.join(dir_train_flowing, 'images')
|
||||
dir_flow_train_labels = os.path.join(dir_train_flowing, 'labels')
|
||||
|
||||
dir_flow_eval_imgs = os.path.join(dir_eval_flowing, 'images')
|
||||
dir_flow_eval_labels = os.path.join(dir_eval_flowing, 'labels')
|
||||
|
||||
configuration()
|
||||
|
||||
else:
|
||||
dir_img, dir_seg = get_dirs_or_files(dir_train)
|
||||
dir_img_val, dir_seg_val = get_dirs_or_files(dir_eval)
|
||||
|
||||
# make first a directory in output for both training and evaluations in order to flow data from these directories.
|
||||
dir_train_flowing = os.path.join(dir_output, 'train')
|
||||
dir_eval_flowing = os.path.join(dir_output, 'eval')
|
||||
|
||||
dir_flow_train_imgs = os.path.join(dir_train_flowing, 'images/')
|
||||
dir_flow_train_labels = os.path.join(dir_train_flowing, 'labels/')
|
||||
|
||||
dir_flow_eval_imgs = os.path.join(dir_eval_flowing, 'images/')
|
||||
dir_flow_eval_labels = os.path.join(dir_eval_flowing, 'labels/')
|
||||
|
||||
if os.path.isdir(dir_train_flowing):
|
||||
os.system('rm -rf ' + dir_train_flowing)
|
||||
os.makedirs(dir_train_flowing)
|
||||
else:
|
||||
os.makedirs(dir_train_flowing)
|
||||
|
||||
if os.path.isdir(dir_eval_flowing):
|
||||
os.system('rm -rf ' + dir_eval_flowing)
|
||||
os.makedirs(dir_eval_flowing)
|
||||
else:
|
||||
os.makedirs(dir_eval_flowing)
|
||||
|
||||
os.mkdir(dir_flow_train_imgs)
|
||||
os.mkdir(dir_flow_train_labels)
|
||||
|
||||
os.mkdir(dir_flow_eval_imgs)
|
||||
os.mkdir(dir_flow_eval_labels)
|
||||
|
||||
# set the gpu configuration
|
||||
configuration()
|
||||
|
||||
imgs_list=np.array(os.listdir(dir_img))
|
||||
segs_list=np.array(os.listdir(dir_seg))
|
||||
|
||||
imgs_list_test=np.array(os.listdir(dir_img_val))
|
||||
segs_list_test=np.array(os.listdir(dir_seg_val))
|
||||
|
||||
# writing patches into a sub-folder in order to be flowed from directory.
|
||||
provide_patches(imgs_list, segs_list, dir_img, dir_seg, dir_flow_train_imgs,
|
||||
dir_flow_train_labels, input_height, input_width, blur_k,
|
||||
blur_aug, padding_white, padding_black, flip_aug, binarization, adding_rgb_background,adding_rgb_foreground, add_red_textlines, channels_shuffling,
|
||||
scaling, shifting, degrading, brightening, scales, degrade_scales, brightness,
|
||||
flip_index,shuffle_indexes, scaling_bluring, scaling_brightness, scaling_binarization,
|
||||
rotation, rotation_not_90, thetha, scaling_flip, task, augmentation=augmentation,
|
||||
patches=patches, dir_img_bin=dir_img_bin,number_of_backgrounds_per_image=number_of_backgrounds_per_image,list_all_possible_background_images=list_all_possible_background_images, dir_rgb_backgrounds=dir_rgb_backgrounds, dir_rgb_foregrounds=dir_rgb_foregrounds,list_all_possible_foreground_rgbs=list_all_possible_foreground_rgbs)
|
||||
|
||||
provide_patches(imgs_list_test, segs_list_test, dir_img_val, dir_seg_val,
|
||||
dir_flow_eval_imgs, dir_flow_eval_labels, input_height, input_width,
|
||||
blur_k, blur_aug, padding_white, padding_black, flip_aug, binarization, adding_rgb_background, adding_rgb_foreground, add_red_textlines, channels_shuffling,
|
||||
scaling, shifting, degrading, brightening, scales, degrade_scales, brightness,
|
||||
flip_index, shuffle_indexes, scaling_bluring, scaling_brightness, scaling_binarization,
|
||||
rotation, rotation_not_90, thetha, scaling_flip, task, augmentation=False, patches=patches,dir_img_bin=dir_img_bin,number_of_backgrounds_per_image=number_of_backgrounds_per_image,list_all_possible_background_images=list_all_possible_background_images, dir_rgb_backgrounds=dir_rgb_backgrounds,dir_rgb_foregrounds=dir_rgb_foregrounds,list_all_possible_foreground_rgbs=list_all_possible_foreground_rgbs )
|
||||
|
||||
if weighted_loss:
|
||||
weights = np.zeros(n_classes)
|
||||
if data_is_provided:
|
||||
for obj in os.listdir(dir_flow_train_labels):
|
||||
try:
|
||||
label_obj = cv2.imread(dir_flow_train_labels + '/' + obj)
|
||||
label_obj_one_hot = get_one_hot(label_obj, label_obj.shape[0], label_obj.shape[1], n_classes)
|
||||
weights += (label_obj_one_hot.sum(axis=0)).sum(axis=0)
|
||||
except:
|
||||
pass
|
||||
else:
|
||||
|
||||
assert dir_seg is not None
|
||||
for obj in os.listdir(dir_seg):
|
||||
try:
|
||||
label_obj = cv2.imread(dir_seg + '/' + obj)
|
||||
label_obj_one_hot = get_one_hot(label_obj, label_obj.shape[0], label_obj.shape[1], n_classes)
|
||||
weights += (label_obj_one_hot.sum(axis=0)).sum(axis=0)
|
||||
except:
|
||||
pass
|
||||
|
||||
weights = 1.00 / weights
|
||||
|
||||
weights = weights / float(np.sum(weights))
|
||||
weights = weights / float(np.min(weights))
|
||||
weights = weights / float(np.sum(weights))
|
||||
|
||||
if continue_training:
|
||||
if backbone_type=='nontransformer':
|
||||
if is_loss_soft_dice and (task == "segmentation" or task == "binarization"):
|
||||
model = load_model(dir_of_start_model, compile=True, custom_objects={'soft_dice_loss': soft_dice_loss})
|
||||
if weighted_loss and (task == "segmentation" or task == "binarization"):
|
||||
model = load_model(dir_of_start_model, compile=True, custom_objects={'loss': weighted_categorical_crossentropy(weights)})
|
||||
if not is_loss_soft_dice and not weighted_loss:
|
||||
model = load_model(dir_of_start_model , compile=True)
|
||||
elif backbone_type=='transformer':
|
||||
if is_loss_soft_dice and (task == "segmentation" or task == "binarization"):
|
||||
model = load_model(dir_of_start_model, compile=True, custom_objects={"PatchEncoder": PatchEncoder, "Patches": Patches,'soft_dice_loss': soft_dice_loss})
|
||||
if weighted_loss and (task == "segmentation" or task == "binarization"):
|
||||
model = load_model(dir_of_start_model, compile=True, custom_objects={'loss': weighted_categorical_crossentropy(weights)})
|
||||
if not is_loss_soft_dice and not weighted_loss:
|
||||
model = load_model(dir_of_start_model , compile=True,custom_objects = {"PatchEncoder": PatchEncoder, "Patches": Patches})
|
||||
else:
|
||||
raise ValueError("backbone_type must be 'nontransformer' or 'transformer'")
|
||||
else:
|
||||
index_start = 0
|
||||
if backbone_type=='nontransformer':
|
||||
model = resnet50_unet(n_classes, input_height, input_width, task, weight_decay, pretraining)
|
||||
elif backbone_type=='transformer':
|
||||
num_patches_x = transformer_num_patches_xy[0]
|
||||
num_patches_y = transformer_num_patches_xy[1]
|
||||
num_patches = num_patches_x * num_patches_y
|
||||
|
||||
if transformer_cnn_first:
|
||||
if input_height != (num_patches_y * transformer_patchsize_y * 32):
|
||||
print("Error: transformer_patchsize_y or transformer_num_patches_xy height value error . input_height should be equal to ( transformer_num_patches_xy height value * transformer_patchsize_y * 32)")
|
||||
sys.exit(1)
|
||||
if input_width != (num_patches_x * transformer_patchsize_x * 32):
|
||||
print("Error: transformer_patchsize_x or transformer_num_patches_xy width value error . input_width should be equal to ( transformer_num_patches_xy width value * transformer_patchsize_x * 32)")
|
||||
sys.exit(1)
|
||||
if (transformer_projection_dim % (transformer_patchsize_y * transformer_patchsize_x)) != 0:
|
||||
print("Error: transformer_projection_dim error. The remainder when parameter transformer_projection_dim is divided by (transformer_patchsize_y*transformer_patchsize_x) should be zero")
|
||||
sys.exit(1)
|
||||
|
||||
|
||||
model = vit_resnet50_unet(n_classes, transformer_patchsize_x, transformer_patchsize_y, num_patches, transformer_mlp_head_units, transformer_layers, transformer_num_heads, transformer_projection_dim, input_height, input_width, task, weight_decay, pretraining)
|
||||
else:
|
||||
if input_height != (num_patches_y * transformer_patchsize_y):
|
||||
print("Error: transformer_patchsize_y or transformer_num_patches_xy height value error . input_height should be equal to ( transformer_num_patches_xy height value * transformer_patchsize_y)")
|
||||
sys.exit(1)
|
||||
if input_width != (num_patches_x * transformer_patchsize_x):
|
||||
print("Error: transformer_patchsize_x or transformer_num_patches_xy width value error . input_width should be equal to ( transformer_num_patches_xy width value * transformer_patchsize_x)")
|
||||
sys.exit(1)
|
||||
if (transformer_projection_dim % (transformer_patchsize_y * transformer_patchsize_x)) != 0:
|
||||
print("Error: transformer_projection_dim error. The remainder when parameter transformer_projection_dim is divided by (transformer_patchsize_y*transformer_patchsize_x) should be zero")
|
||||
sys.exit(1)
|
||||
model = vit_resnet50_unet_transformer_before_cnn(n_classes, transformer_patchsize_x, transformer_patchsize_y, num_patches, transformer_mlp_head_units, transformer_layers, transformer_num_heads, transformer_projection_dim, input_height, input_width, task, weight_decay, pretraining)
|
||||
|
||||
assert model is not None
|
||||
#if you want to see the model structure just uncomment model summary.
|
||||
model.summary()
|
||||
|
||||
|
||||
if task == "segmentation" or task == "binarization":
|
||||
if not is_loss_soft_dice and not weighted_loss:
|
||||
model.compile(loss='categorical_crossentropy',
|
||||
optimizer=Adam(learning_rate=learning_rate), metrics=['accuracy'])
|
||||
if is_loss_soft_dice:
|
||||
model.compile(loss=soft_dice_loss,
|
||||
optimizer=Adam(learning_rate=learning_rate), metrics=['accuracy'])
|
||||
if weighted_loss:
|
||||
model.compile(loss=weighted_categorical_crossentropy(weights),
|
||||
optimizer=Adam(learning_rate=learning_rate), metrics=['accuracy'])
|
||||
elif task == "enhancement":
|
||||
model.compile(loss='mean_squared_error',
|
||||
optimizer=Adam(learning_rate=learning_rate), metrics=['accuracy'])
|
||||
|
||||
|
||||
# generating train and evaluation data
|
||||
train_gen = data_gen(dir_flow_train_imgs, dir_flow_train_labels, batch_size=n_batch,
|
||||
input_height=input_height, input_width=input_width, n_classes=n_classes, task=task)
|
||||
val_gen = data_gen(dir_flow_eval_imgs, dir_flow_eval_labels, batch_size=n_batch,
|
||||
input_height=input_height, input_width=input_width, n_classes=n_classes, task=task)
|
||||
|
||||
##img_validation_patches = os.listdir(dir_flow_eval_imgs)
|
||||
##score_best=[]
|
||||
##score_best.append(0)
|
||||
|
||||
save_weights_callback = SaveWeightsAfterSteps(save_interval, dir_output, _config) if save_interval else None
|
||||
|
||||
for i in tqdm(range(index_start, n_epochs + index_start)):
|
||||
if save_interval:
|
||||
model.fit(
|
||||
train_gen,
|
||||
steps_per_epoch=int(len(os.listdir(dir_flow_train_imgs)) / n_batch) - 1,
|
||||
validation_data=val_gen,
|
||||
validation_steps=1,
|
||||
epochs=1, callbacks=[save_weights_callback])
|
||||
else:
|
||||
model.fit(
|
||||
train_gen,
|
||||
steps_per_epoch=int(len(os.listdir(dir_flow_train_imgs)) / n_batch) - 1,
|
||||
validation_data=val_gen,
|
||||
validation_steps=1,
|
||||
epochs=1)
|
||||
|
||||
model.save(os.path.join(dir_output,'model_'+str(i)))
|
||||
|
||||
with open(os.path.join(os.path.join(dir_output,'model_'+str(i)),"config.json"), "w") as fp:
|
||||
json.dump(_config, fp) # encode dict into JSON
|
||||
|
||||
#os.system('rm -rf '+dir_train_flowing)
|
||||
#os.system('rm -rf '+dir_eval_flowing)
|
||||
|
||||
#model.save(dir_output+'/'+'model'+'.h5')
|
||||
|
||||
elif task=="cnn-rnn-ocr":
|
||||
dir_img, dir_lab = get_dirs_or_files(dir_train)
|
||||
|
||||
with open(characters_txt_file, 'r') as char_txt_f:
|
||||
characters = json.load(char_txt_f)
|
||||
|
||||
AUTOTUNE = tf.data.AUTOTUNE
|
||||
|
||||
# Mapping characters to integers.
|
||||
char_to_num = StringLookup(vocabulary=list(characters), mask_token=None)
|
||||
|
||||
# Mapping integers back to original characters.
|
||||
##num_to_char = StringLookup(
|
||||
##vocabulary=char_to_num.get_vocabulary(), mask_token=None, invert=True
|
||||
##)
|
||||
|
||||
padding_token = len(characters) + 5
|
||||
ls_files_images = os.listdir(dir_img)
|
||||
|
||||
n_classes = len(char_to_num.get_vocabulary()) + 2
|
||||
|
||||
if continue_training:
|
||||
model = load_model(dir_of_start_model)
|
||||
else:
|
||||
index_start = 0
|
||||
model = cnn_rnn_ocr_model(image_height=input_height, image_width=input_width, n_classes=n_classes, max_seq=max_len)
|
||||
|
||||
print(model.summary())
|
||||
|
||||
aug_multip = return_multiplier_based_on_augmnentations(augmentation, color_padding_rotation, rotation_not_90, blur_aug, degrading, bin_deg,
|
||||
brightening, padding_white, adding_rgb_foreground, adding_rgb_background, binarization,
|
||||
image_inversion, channels_shuffling, add_red_textlines, white_noise_strap, textline_skewing, textline_skewing_bin, textline_left_in_depth, textline_left_in_depth_bin, textline_right_in_depth, textline_right_in_depth_bin, textline_up_in_depth, textline_up_in_depth_bin, textline_down_in_depth, textline_down_in_depth_bin, pepper_bin_aug, pepper_aug, degrade_scales, number_of_backgrounds_per_image, thetha, thetha_padd, brightness, padd_colors, shuffle_indexes, pepper_indexes, skewing_amplitudes, blur_k, white_padds)
|
||||
|
||||
len_dataset = aug_multip*len(ls_files_images)
|
||||
|
||||
train_ds = data_gen_ocr(padding_token, n_batch, input_height, input_width, max_len, dir_train, ls_files_images,
|
||||
augmentation, color_padding_rotation, rotation_not_90, blur_aug, degrading, bin_deg, brightening, padding_white,
|
||||
adding_rgb_foreground, adding_rgb_background, binarization, image_inversion, channels_shuffling, add_red_textlines, white_noise_strap,
|
||||
textline_skewing, textline_skewing_bin, textline_left_in_depth, textline_left_in_depth_bin, textline_right_in_depth,
|
||||
textline_right_in_depth_bin, textline_up_in_depth, textline_up_in_depth_bin, textline_down_in_depth, textline_down_in_depth_bin,
|
||||
pepper_bin_aug, pepper_aug, degrade_scales, number_of_backgrounds_per_image, thetha, thetha_padd, brightness, padd_colors,
|
||||
shuffle_indexes, pepper_indexes, skewing_amplitudes, blur_k, char_to_num, list_all_possible_background_images, list_all_possible_foreground_rgbs,
|
||||
dir_rgb_backgrounds, dir_rgb_foregrounds, white_padds, dir_img_bin)
|
||||
|
||||
initial_learning_rate = 1e-4
|
||||
decay_steps = int (n_epochs * ( len_dataset / n_batch ))
|
||||
alpha = 0.01
|
||||
lr_schedule = 1e-4#tf.keras.optimizers.schedules.CosineDecay(initial_learning_rate, decay_steps, alpha)
|
||||
|
||||
opt = tf.keras.optimizers.Adam(learning_rate=learning_rate)#1e-4)#(lr_schedule)
|
||||
model.compile(optimizer=opt)
|
||||
|
||||
save_weights_callback = SaveWeightsAfterSteps(save_interval, dir_output, _config) if save_interval else None
|
||||
|
||||
for i in tqdm(range(index_start, n_epochs + index_start)):
|
||||
if save_interval:
|
||||
model.fit(
|
||||
train_ds,
|
||||
steps_per_epoch=len_dataset / n_batch,
|
||||
epochs=1,
|
||||
callbacks=[save_weights_callback]
|
||||
)
|
||||
else:
|
||||
model.fit(
|
||||
train_ds,
|
||||
steps_per_epoch=len_dataset / n_batch,
|
||||
epochs=1
|
||||
)
|
||||
|
||||
if i >=0:
|
||||
model.save( os.path.join(dir_output,'model_'+str(i) ))
|
||||
with open(os.path.join(os.path.join(dir_output,'model_'+str(i)),"config.json"), "w") as fp:
|
||||
json.dump(_config, fp) # encode dict into JSON
|
||||
|
||||
elif task=='classification':
|
||||
configuration()
|
||||
model = resnet50_classifier(n_classes, input_height, input_width, weight_decay, pretraining)
|
||||
|
||||
opt_adam = Adam(learning_rate=0.001)
|
||||
model.compile(loss='categorical_crossentropy',
|
||||
optimizer = opt_adam,metrics=['accuracy'])
|
||||
|
||||
|
||||
list_classes = list(classification_classes_name.values())
|
||||
testX, testY = generate_data_from_folder_evaluation(dir_eval, input_height, input_width, n_classes, list_classes)
|
||||
|
||||
y_tot=np.zeros((testX.shape[0],n_classes))
|
||||
|
||||
score_best= [0]
|
||||
|
||||
num_rows = return_number_of_total_training_data(dir_train)
|
||||
weights=[]
|
||||
|
||||
for i in range(n_epochs):
|
||||
history = model.fit( generate_data_from_folder_training(dir_train, n_batch , input_height, input_width, n_classes, list_classes), steps_per_epoch=num_rows / n_batch, verbose=1)#,class_weight=weights)
|
||||
|
||||
y_pr_class = []
|
||||
for jj in range(testY.shape[0]):
|
||||
y_pr=model.predict(testX[jj,:,:,:].reshape(1,input_height,input_width,3), verbose=0)
|
||||
y_pr_ind= np.argmax(y_pr,axis=1)
|
||||
y_pr_class.append(y_pr_ind)
|
||||
|
||||
y_pr_class = np.array(y_pr_class)
|
||||
f1score=f1_score(np.argmax(testY,axis=1), y_pr_class, average='macro')
|
||||
print(i,f1score)
|
||||
|
||||
if f1score>score_best[0]:
|
||||
score_best[0]=f1score
|
||||
model.save(os.path.join(dir_output,'model_best'))
|
||||
|
||||
if f1score > f1_threshold_classification:
|
||||
weights.append(model.get_weights() )
|
||||
|
||||
|
||||
if len(weights) >= 1:
|
||||
new_weights=list()
|
||||
for weights_list_tuple in zip(*weights):
|
||||
new_weights.append( [np.array(weights_).mean(axis=0) for weights_ in zip(*weights_list_tuple)] )
|
||||
|
||||
new_weights = [np.array(x) for x in new_weights]
|
||||
model_weight_averaged=tf.keras.models.clone_model(model)
|
||||
model_weight_averaged.set_weights(new_weights)
|
||||
|
||||
model_weight_averaged.save(os.path.join(dir_output,'model_ens_avg'))
|
||||
with open(os.path.join( os.path.join(dir_output,'model_ens_avg'), "config.json"), "w") as fp:
|
||||
json.dump(_config, fp) # encode dict into JSON
|
||||
|
||||
with open(os.path.join( os.path.join(dir_output,'model_best'), "config.json"), "w") as fp:
|
||||
json.dump(_config, fp) # encode dict into JSON
|
||||
|
||||
elif task=='reading_order':
|
||||
configuration()
|
||||
model = machine_based_reading_order_model(n_classes,input_height,input_width,weight_decay,pretraining)
|
||||
|
||||
dir_flow_train_imgs = os.path.join(dir_train, 'images')
|
||||
dir_flow_train_labels = os.path.join(dir_train, 'labels')
|
||||
|
||||
classes = os.listdir(dir_flow_train_labels)
|
||||
if augmentation:
|
||||
num_rows = len(classes)*(len(thetha) + 1)
|
||||
else:
|
||||
num_rows = len(classes)
|
||||
#ls_test = os.listdir(dir_flow_train_labels)
|
||||
|
||||
#f1score_tot = [0]
|
||||
indexer_start = 0
|
||||
# opt = SGD(learning_rate=0.01, momentum=0.9)
|
||||
opt_adam = tf.keras.optimizers.Adam(learning_rate=0.0001)
|
||||
model.compile(loss="binary_crossentropy",
|
||||
optimizer = opt_adam,metrics=['accuracy'])
|
||||
|
||||
save_weights_callback = SaveWeightsAfterSteps(save_interval, dir_output, _config) if save_interval else None
|
||||
|
||||
for i in range(n_epochs):
|
||||
if save_interval:
|
||||
history = model.fit(generate_arrays_from_folder_reading_order(dir_flow_train_labels, dir_flow_train_imgs, n_batch, input_height, input_width, n_classes, thetha, augmentation), steps_per_epoch=num_rows / n_batch, verbose=1, callbacks=[save_weights_callback])
|
||||
else:
|
||||
history = model.fit(generate_arrays_from_folder_reading_order(dir_flow_train_labels, dir_flow_train_imgs, n_batch, input_height, input_width, n_classes, thetha, augmentation), steps_per_epoch=num_rows / n_batch, verbose=1)
|
||||
model.save( os.path.join(dir_output,'model_'+str(i+indexer_start) ))
|
||||
|
||||
with open(os.path.join(os.path.join(dir_output,'model_'+str(i)),"config.json"), "w") as fp:
|
||||
json.dump(_config, fp) # encode dict into JSON
|
||||
'''
|
||||
if f1score>f1score_tot[0]:
|
||||
f1score_tot[0] = f1score
|
||||
model_dir = os.path.join(dir_out,'model_best')
|
||||
model.save(model_dir)
|
||||
'''
|
||||
|
||||
|
||||
File diff suppressed because it is too large
Load diff
|
|
@ -1,136 +0,0 @@
|
|||
import sys
|
||||
from glob import glob
|
||||
from os import environ, devnull
|
||||
from os.path import join
|
||||
from warnings import catch_warnings, simplefilter
|
||||
import os
|
||||
|
||||
import numpy as np
|
||||
from PIL import Image
|
||||
import cv2
|
||||
environ['TF_CPP_MIN_LOG_LEVEL'] = '3'
|
||||
stderr = sys.stderr
|
||||
sys.stderr = open(devnull, 'w')
|
||||
import tensorflow as tf
|
||||
from tensorflow.keras.models import load_model
|
||||
from tensorflow.python.keras import backend as tensorflow_backend
|
||||
sys.stderr = stderr
|
||||
from tensorflow.keras import layers
|
||||
import tensorflow.keras.losses
|
||||
from tensorflow.keras.layers import *
|
||||
import click
|
||||
import logging
|
||||
|
||||
|
||||
class Patches(layers.Layer):
|
||||
def __init__(self, patch_size_x, patch_size_y):
|
||||
super(Patches, self).__init__()
|
||||
self.patch_size_x = patch_size_x
|
||||
self.patch_size_y = patch_size_y
|
||||
|
||||
def call(self, images):
|
||||
#print(tf.shape(images)[1],'images')
|
||||
#print(self.patch_size,'self.patch_size')
|
||||
batch_size = tf.shape(images)[0]
|
||||
patches = tf.image.extract_patches(
|
||||
images=images,
|
||||
sizes=[1, self.patch_size_y, self.patch_size_x, 1],
|
||||
strides=[1, self.patch_size_y, self.patch_size_x, 1],
|
||||
rates=[1, 1, 1, 1],
|
||||
padding="VALID",
|
||||
)
|
||||
#patch_dims = patches.shape[-1]
|
||||
patch_dims = tf.shape(patches)[-1]
|
||||
patches = tf.reshape(patches, [batch_size, -1, patch_dims])
|
||||
return patches
|
||||
def get_config(self):
|
||||
|
||||
config = super().get_config().copy()
|
||||
config.update({
|
||||
'patch_size_x': self.patch_size_x,
|
||||
'patch_size_y': self.patch_size_y,
|
||||
})
|
||||
return config
|
||||
|
||||
|
||||
|
||||
class PatchEncoder(layers.Layer):
|
||||
def __init__(self, **kwargs):
|
||||
super(PatchEncoder, self).__init__()
|
||||
self.num_patches = num_patches
|
||||
self.projection = layers.Dense(units=projection_dim)
|
||||
self.position_embedding = layers.Embedding(
|
||||
input_dim=num_patches, output_dim=projection_dim
|
||||
)
|
||||
|
||||
def call(self, patch):
|
||||
positions = tf.range(start=0, limit=self.num_patches, delta=1)
|
||||
encoded = self.projection(patch) + self.position_embedding(positions)
|
||||
return encoded
|
||||
def get_config(self):
|
||||
|
||||
config = super().get_config().copy()
|
||||
config.update({
|
||||
'num_patches': self.num_patches,
|
||||
'projection': self.projection,
|
||||
'position_embedding': self.position_embedding,
|
||||
})
|
||||
return config
|
||||
|
||||
|
||||
def start_new_session():
|
||||
###config = tf.compat.v1.ConfigProto()
|
||||
###config.gpu_options.allow_growth = True
|
||||
|
||||
###self.session = tf.compat.v1.Session(config=config) # tf.InteractiveSession()
|
||||
###tensorflow_backend.set_session(self.session)
|
||||
|
||||
config = tf.compat.v1.ConfigProto()
|
||||
config.gpu_options.allow_growth = True
|
||||
|
||||
session = tf.compat.v1.Session(config=config) # tf.InteractiveSession()
|
||||
tensorflow_backend.set_session(session)
|
||||
return session
|
||||
|
||||
def run_ensembling(dir_models, out):
|
||||
ls_models = os.listdir(dir_models)
|
||||
|
||||
|
||||
weights=[]
|
||||
|
||||
for model_name in ls_models:
|
||||
model = load_model(os.path.join(dir_models,model_name) , compile=False, custom_objects={'PatchEncoder':PatchEncoder, 'Patches': Patches})
|
||||
weights.append(model.get_weights())
|
||||
|
||||
new_weights = list()
|
||||
|
||||
for weights_list_tuple in zip(*weights):
|
||||
new_weights.append(
|
||||
[np.array(weights_).mean(axis=0)\
|
||||
for weights_ in zip(*weights_list_tuple)])
|
||||
|
||||
|
||||
|
||||
new_weights = [np.array(x) for x in new_weights]
|
||||
|
||||
model.set_weights(new_weights)
|
||||
model.save(out)
|
||||
os.system('cp '+os.path.join(os.path.join(dir_models,model_name) , "config.json ")+out)
|
||||
|
||||
@click.command()
|
||||
@click.option(
|
||||
"--dir_models",
|
||||
"-dm",
|
||||
help="directory of models",
|
||||
type=click.Path(exists=True, file_okay=False),
|
||||
)
|
||||
@click.option(
|
||||
"--out",
|
||||
"-o",
|
||||
help="output directory where ensembled model will be written.",
|
||||
type=click.Path(exists=False, file_okay=False),
|
||||
)
|
||||
|
||||
def main(dir_models, out):
|
||||
run_ensembling(dir_models, out)
|
||||
|
||||
File diff suppressed because it is too large
Load diff
|
|
@ -1,15 +1,7 @@
|
|||
from typing import Sequence, Union
|
||||
from numbers import Number
|
||||
from functools import partial
|
||||
import itertools
|
||||
|
||||
import cv2
|
||||
import numpy as np
|
||||
from scipy.sparse.csgraph import minimum_spanning_tree
|
||||
from shapely.geometry import Polygon, LineString
|
||||
from shapely.geometry.polygon import orient
|
||||
from shapely import set_precision
|
||||
from shapely.ops import unary_union, nearest_points
|
||||
from shapely import geometry
|
||||
|
||||
from .rotate import rotate_image, rotation_image_new
|
||||
|
||||
|
|
@ -36,31 +28,38 @@ def find_contours_mean_y_diff(contours_main):
|
|||
return np.mean(np.diff(np.sort(np.array(cy_main))))
|
||||
|
||||
def get_text_region_boxes_by_given_contours(contours):
|
||||
return [cv2.boundingRect(contour)
|
||||
for contour in contours]
|
||||
boxes = []
|
||||
contours_new = []
|
||||
for jj in range(len(contours)):
|
||||
box = cv2.boundingRect(contours[jj])
|
||||
boxes.append(box)
|
||||
contours_new.append(contours[jj])
|
||||
|
||||
def filter_contours_area_of_image(image, contours, hierarchy, max_area=1.0, min_area=0.0, dilate=0):
|
||||
return boxes, contours_new
|
||||
|
||||
def filter_contours_area_of_image(image, contours, hierarchy, max_area, min_area):
|
||||
found_polygons_early = []
|
||||
for jv, contour in enumerate(contours):
|
||||
if len(contour) < 3: # A polygon cannot have less than 3 points
|
||||
for jv,c in enumerate(contours):
|
||||
if len(c) < 3: # A polygon cannot have less than 3 points
|
||||
continue
|
||||
|
||||
polygon = contour2polygon(contour, dilate=dilate)
|
||||
polygon = geometry.Polygon([point[0] for point in c])
|
||||
area = polygon.area
|
||||
if (area >= min_area * np.prod(image.shape[:2]) and
|
||||
area <= max_area * np.prod(image.shape[:2]) and
|
||||
hierarchy[0][jv][3] == -1):
|
||||
found_polygons_early.append(polygon2contour(polygon))
|
||||
found_polygons_early.append(np.array([[point]
|
||||
for point in polygon.exterior.coords], dtype=np.uint))
|
||||
return found_polygons_early
|
||||
|
||||
def filter_contours_area_of_image_tables(image, contours, hierarchy, max_area=1.0, min_area=0.0, dilate=0):
|
||||
def filter_contours_area_of_image_tables(image, contours, hierarchy, max_area, min_area):
|
||||
found_polygons_early = []
|
||||
for jv, contour in enumerate(contours):
|
||||
if len(contour) < 3: # A polygon cannot have less than 3 points
|
||||
for jv,c in enumerate(contours):
|
||||
if len(c) < 3: # A polygon cannot have less than 3 points
|
||||
continue
|
||||
|
||||
polygon = contour2polygon(contour, dilate=dilate)
|
||||
# area = cv2.contourArea(contour)
|
||||
polygon = geometry.Polygon([point[0] for point in c])
|
||||
# area = cv2.contourArea(c)
|
||||
area = polygon.area
|
||||
##print(np.prod(thresh.shape[:2]))
|
||||
# Check that polygon has area greater than minimal area
|
||||
|
|
@ -69,41 +68,66 @@ def filter_contours_area_of_image_tables(image, contours, hierarchy, max_area=1.
|
|||
area <= max_area * np.prod(image.shape[:2]) and
|
||||
# hierarchy[0][jv][3]==-1
|
||||
True):
|
||||
# print(contour[0][0][1])
|
||||
found_polygons_early.append(polygon2contour(polygon))
|
||||
# print(c[0][0][1])
|
||||
found_polygons_early.append(np.array([[point]
|
||||
for point in polygon.exterior.coords], dtype=np.int32))
|
||||
return found_polygons_early
|
||||
|
||||
def find_center_of_contours(contours):
|
||||
moments = [cv2.moments(contour) for contour in contours]
|
||||
cx = [feat["m10"] / (feat["m00"] + 1e-32)
|
||||
for feat in moments]
|
||||
cy = [feat["m01"] / (feat["m00"] + 1e-32)
|
||||
for feat in moments]
|
||||
return cx, cy
|
||||
def find_new_features_of_contours(contours_main):
|
||||
areas_main = np.array([cv2.contourArea(contours_main[j])
|
||||
for j in range(len(contours_main))])
|
||||
M_main = [cv2.moments(contours_main[j])
|
||||
for j in range(len(contours_main))]
|
||||
cx_main = [(M_main[j]["m10"] / (M_main[j]["m00"] + 1e-32))
|
||||
for j in range(len(M_main))]
|
||||
cy_main = [(M_main[j]["m01"] / (M_main[j]["m00"] + 1e-32))
|
||||
for j in range(len(M_main))]
|
||||
try:
|
||||
x_min_main = np.array([np.min(contours_main[j][:, 0, 0])
|
||||
for j in range(len(contours_main))])
|
||||
argmin_x_main = np.array([np.argmin(contours_main[j][:, 0, 0])
|
||||
for j in range(len(contours_main))])
|
||||
x_min_from_argmin = np.array([contours_main[j][argmin_x_main[j], 0, 0]
|
||||
for j in range(len(contours_main))])
|
||||
y_corr_x_min_from_argmin = np.array([contours_main[j][argmin_x_main[j], 0, 1]
|
||||
for j in range(len(contours_main))])
|
||||
x_max_main = np.array([np.max(contours_main[j][:, 0, 0])
|
||||
for j in range(len(contours_main))])
|
||||
y_min_main = np.array([np.min(contours_main[j][:, 0, 1])
|
||||
for j in range(len(contours_main))])
|
||||
y_max_main = np.array([np.max(contours_main[j][:, 0, 1])
|
||||
for j in range(len(contours_main))])
|
||||
except:
|
||||
x_min_main = np.array([np.min(contours_main[j][:, 0])
|
||||
for j in range(len(contours_main))])
|
||||
argmin_x_main = np.array([np.argmin(contours_main[j][:, 0])
|
||||
for j in range(len(contours_main))])
|
||||
x_min_from_argmin = np.array([contours_main[j][argmin_x_main[j], 0]
|
||||
for j in range(len(contours_main))])
|
||||
y_corr_x_min_from_argmin = np.array([contours_main[j][argmin_x_main[j], 1]
|
||||
for j in range(len(contours_main))])
|
||||
x_max_main = np.array([np.max(contours_main[j][:, 0])
|
||||
for j in range(len(contours_main))])
|
||||
y_min_main = np.array([np.min(contours_main[j][:, 1])
|
||||
for j in range(len(contours_main))])
|
||||
y_max_main = np.array([np.max(contours_main[j][:, 1])
|
||||
for j in range(len(contours_main))])
|
||||
# dis_x=np.abs(x_max_main-x_min_main)
|
||||
|
||||
def find_new_features_of_contours(contours):
|
||||
# areas = np.array([cv2.contourArea(contour) for contour in contours])
|
||||
cx, cy = find_center_of_contours(contours)
|
||||
slice_x = np.index_exp[:, 0, 0]
|
||||
slice_y = np.index_exp[:, 0, 1]
|
||||
if any(contour.ndim < 3 for contour in contours):
|
||||
slice_x = np.index_exp[:, 0]
|
||||
slice_y = np.index_exp[:, 1]
|
||||
x_min = np.array([np.min(contour[slice_x]) for contour in contours])
|
||||
x_max = np.array([np.max(contour[slice_x]) for contour in contours])
|
||||
y_min = np.array([np.min(contour[slice_y]) for contour in contours])
|
||||
y_max = np.array([np.max(contour[slice_y]) for contour in contours])
|
||||
# dis_x=np.abs(x_max-x_min)
|
||||
y_corr_x_min = np.array([contour[np.argmin(contour[slice_x])][slice_y[1:]]
|
||||
for contour in contours])
|
||||
return cx_main, cy_main, x_min_main, x_max_main, y_min_main, y_max_main, y_corr_x_min_from_argmin
|
||||
|
||||
return cx, cy, x_min, x_max, y_min, y_max, y_corr_x_min
|
||||
def find_features_of_contours(contours_main):
|
||||
areas_main=np.array([cv2.contourArea(contours_main[j]) for j in range(len(contours_main))])
|
||||
M_main=[cv2.moments(contours_main[j]) for j in range(len(contours_main))]
|
||||
cx_main=[(M_main[j]['m10']/(M_main[j]['m00']+1e-32)) for j in range(len(M_main))]
|
||||
cy_main=[(M_main[j]['m01']/(M_main[j]['m00']+1e-32)) for j in range(len(M_main))]
|
||||
x_min_main=np.array([np.min(contours_main[j][:,0,0]) for j in range(len(contours_main))])
|
||||
x_max_main=np.array([np.max(contours_main[j][:,0,0]) for j in range(len(contours_main))])
|
||||
|
||||
def find_features_of_contours(contours):
|
||||
y_min = np.array([np.min(contour[:,0,1]) for contour in contours])
|
||||
y_max = np.array([np.max(contour[:,0,1]) for contour in contours])
|
||||
y_min_main=np.array([np.min(contours_main[j][:,0,1]) for j in range(len(contours_main))])
|
||||
y_max_main=np.array([np.max(contours_main[j][:,0,1]) for j in range(len(contours_main))])
|
||||
|
||||
return y_min, y_max
|
||||
return y_min_main, y_max_main
|
||||
|
||||
def return_parent_contours(contours, hierarchy):
|
||||
contours_parent = [contours[i]
|
||||
|
|
@ -111,13 +135,16 @@ def return_parent_contours(contours, hierarchy):
|
|||
if hierarchy[0][i][3] == -1]
|
||||
return contours_parent
|
||||
|
||||
def return_contours_of_interested_region(region_pre_p, label, min_area=0.0002):
|
||||
def return_contours_of_interested_region(region_pre_p, pixel, min_area=0.0002):
|
||||
# pixels of images are identified by 5
|
||||
if region_pre_p.ndim == 3:
|
||||
cnts_images = (region_pre_p[:, :, 0] == label) * 1
|
||||
if len(region_pre_p.shape) == 3:
|
||||
cnts_images = (region_pre_p[:, :, 0] == pixel) * 1
|
||||
else:
|
||||
cnts_images = (region_pre_p[:, :] == label) * 1
|
||||
_, thresh = cv2.threshold(cnts_images.astype(np.uint8), 0, 255, 0)
|
||||
cnts_images = (region_pre_p[:, :] == pixel) * 1
|
||||
cnts_images = cnts_images.astype(np.uint8)
|
||||
cnts_images = np.repeat(cnts_images[:, :, np.newaxis], 3, axis=2)
|
||||
imgray = cv2.cvtColor(cnts_images, cv2.COLOR_BGR2GRAY)
|
||||
ret, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
|
||||
contours_imgs, hierarchy = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
contours_imgs = return_parent_contours(contours_imgs, hierarchy)
|
||||
|
|
@ -126,11 +153,13 @@ def return_contours_of_interested_region(region_pre_p, label, min_area=0.0002):
|
|||
return contours_imgs
|
||||
|
||||
def do_work_of_contours_in_image(contour, index_r_con, img, slope_first):
|
||||
img_copy = np.zeros(img.shape[:2], dtype=np.uint8)
|
||||
img_copy = cv2.fillPoly(img_copy, pts=[contour], color=1)
|
||||
img_copy = np.zeros(img.shape)
|
||||
img_copy = cv2.fillPoly(img_copy, pts=[contour], color=(1, 1, 1))
|
||||
|
||||
img_copy = rotation_image_new(img_copy, -slope_first)
|
||||
_, thresh = cv2.threshold(img_copy, 0, 255, 0)
|
||||
img_copy = img_copy.astype(np.uint8)
|
||||
imgray = cv2.cvtColor(img_copy, cv2.COLOR_BGR2GRAY)
|
||||
ret, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
|
||||
cont_int, _ = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
|
||||
|
|
@ -153,8 +182,8 @@ def get_textregion_contours_in_org_image(cnts, img, slope_first):
|
|||
cnts_org = []
|
||||
# print(cnts,'cnts')
|
||||
for i in range(len(cnts)):
|
||||
img_copy = np.zeros(img.shape[:2], dtype=np.uint8)
|
||||
img_copy = cv2.fillPoly(img_copy, pts=[cnts[i]], color=1)
|
||||
img_copy = np.zeros(img.shape)
|
||||
img_copy = cv2.fillPoly(img_copy, pts=[cnts[i]], color=(1, 1, 1))
|
||||
|
||||
# plt.imshow(img_copy)
|
||||
# plt.show()
|
||||
|
|
@ -165,7 +194,9 @@ def get_textregion_contours_in_org_image(cnts, img, slope_first):
|
|||
# plt.imshow(img_copy)
|
||||
# plt.show()
|
||||
|
||||
_, thresh = cv2.threshold(img_copy, 0, 255, 0)
|
||||
img_copy = img_copy.astype(np.uint8)
|
||||
imgray = cv2.cvtColor(img_copy, cv2.COLOR_BGR2GRAY)
|
||||
ret, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
|
||||
cont_int, _ = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
cont_int[0][:, 0, 0] = cont_int[0][:, 0, 0] + np.abs(img_copy.shape[1] - img.shape[1])
|
||||
|
|
@ -182,11 +213,12 @@ def get_textregion_contours_in_org_image_light_old(cnts, img, slope_first):
|
|||
interpolation=cv2.INTER_NEAREST)
|
||||
cnts_org = []
|
||||
for cnt in cnts:
|
||||
img_copy = np.zeros(img.shape[:2], dtype=np.uint8)
|
||||
img_copy = cv2.fillPoly(img_copy, pts=[cnt // zoom], color=1)
|
||||
img_copy = np.zeros(img.shape)
|
||||
img_copy = cv2.fillPoly(img_copy, pts=[(cnt / zoom).astype(int)], color=(1, 1, 1))
|
||||
|
||||
img_copy = rotation_image_new(img_copy, -slope_first).astype(np.uint8)
|
||||
_, thresh = cv2.threshold(img_copy, 0, 255, 0)
|
||||
imgray = cv2.cvtColor(img_copy, cv2.COLOR_BGR2GRAY)
|
||||
ret, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
|
||||
cont_int, _ = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
cont_int[0][:, 0, 0] = cont_int[0][:, 0, 0] + np.abs(img_copy.shape[1] - img.shape[1])
|
||||
|
|
@ -196,44 +228,50 @@ def get_textregion_contours_in_org_image_light_old(cnts, img, slope_first):
|
|||
return cnts_org
|
||||
|
||||
def do_back_rotation_and_get_cnt_back(contour_par, index_r_con, img, slope_first, confidence_matrix):
|
||||
img_copy = np.zeros(img.shape[:2], dtype=np.uint8)
|
||||
img_copy = cv2.fillPoly(img_copy, pts=[contour_par], color=1)
|
||||
confidence_matrix_mapped_with_contour = confidence_matrix * img_copy
|
||||
confidence_contour = np.sum(confidence_matrix_mapped_with_contour) / float(np.sum(img_copy))
|
||||
img_copy = np.zeros(img.shape)
|
||||
img_copy = cv2.fillPoly(img_copy, pts=[contour_par], color=(1, 1, 1))
|
||||
|
||||
confidence_matrix_mapped_with_contour = confidence_matrix * img_copy[:,:,0]
|
||||
confidence_contour = np.sum(confidence_matrix_mapped_with_contour) / float(np.sum(img_copy[:,:,0]))
|
||||
|
||||
img_copy = rotation_image_new(img_copy, -slope_first).astype(np.uint8)
|
||||
_, thresh = cv2.threshold(img_copy, 0, 255, 0)
|
||||
imgray = cv2.cvtColor(img_copy, cv2.COLOR_BGR2GRAY)
|
||||
ret, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
|
||||
cont_int, _ = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
if len(cont_int)==0:
|
||||
cont_int = [contour_par]
|
||||
confidence_contour = 0
|
||||
else:
|
||||
cont_int[0][:, 0, 0] = cont_int[0][:, 0, 0] + np.abs(img_copy.shape[1] - img.shape[1])
|
||||
cont_int[0][:, 0, 1] = cont_int[0][:, 0, 1] + np.abs(img_copy.shape[0] - img.shape[0])
|
||||
cont_int[0][:, 0, 0] = cont_int[0][:, 0, 0] + np.abs(img_copy.shape[1] - img.shape[1])
|
||||
cont_int[0][:, 0, 1] = cont_int[0][:, 0, 1] + np.abs(img_copy.shape[0] - img.shape[0])
|
||||
# print(np.shape(cont_int[0]))
|
||||
return cont_int[0], index_r_con, confidence_contour
|
||||
|
||||
def get_textregion_contours_in_org_image_light(cnts, img, confidence_matrix):
|
||||
def get_textregion_contours_in_org_image_light(cnts, img, slope_first, confidence_matrix, map=map):
|
||||
if not len(cnts):
|
||||
return []
|
||||
return [], []
|
||||
|
||||
confidence_matrix = cv2.resize(confidence_matrix, (int(img.shape[1]/6), int(img.shape[0]/6)), interpolation=cv2.INTER_NEAREST)
|
||||
img = cv2.resize(img, (int(img.shape[1]/6), int(img.shape[0]/6)), interpolation=cv2.INTER_NEAREST)
|
||||
##cnts = list( (np.array(cnts)/2).astype(np.int16) )
|
||||
#cnts = cnts/2
|
||||
cnts = [(i/6).astype(int) for i in cnts]
|
||||
results = map(partial(do_back_rotation_and_get_cnt_back,
|
||||
img=img,
|
||||
slope_first=slope_first,
|
||||
confidence_matrix=confidence_matrix,
|
||||
),
|
||||
cnts, range(len(cnts)))
|
||||
contours, indexes, conf_contours = tuple(zip(*results))
|
||||
return [i*6 for i in contours], list(conf_contours)
|
||||
|
||||
confidence_matrix = cv2.resize(confidence_matrix,
|
||||
(img.shape[1] // 6, img.shape[0] // 6),
|
||||
interpolation=cv2.INTER_NEAREST)
|
||||
confs = []
|
||||
for cnt in cnts:
|
||||
cnt_mask = np.zeros(confidence_matrix.shape)
|
||||
cnt_mask = cv2.fillPoly(cnt_mask, pts=[cnt // 6], color=1.0)
|
||||
confs.append(np.sum(confidence_matrix * cnt_mask) / np.sum(cnt_mask))
|
||||
return confs
|
||||
|
||||
def return_contours_of_interested_textline(region_pre_p, label):
|
||||
def return_contours_of_interested_textline(region_pre_p, pixel):
|
||||
# pixels of images are identified by 5
|
||||
if region_pre_p.ndim == 3:
|
||||
cnts_images = (region_pre_p[:, :, 0] == label) * 1
|
||||
if len(region_pre_p.shape) == 3:
|
||||
cnts_images = (region_pre_p[:, :, 0] == pixel) * 1
|
||||
else:
|
||||
cnts_images = (region_pre_p[:, :] == label) * 1
|
||||
_, thresh = cv2.threshold(cnts_images.astype(np.uint8), 0, 255, 0)
|
||||
cnts_images = (region_pre_p[:, :] == pixel) * 1
|
||||
cnts_images = cnts_images.astype(np.uint8)
|
||||
cnts_images = np.repeat(cnts_images[:, :, np.newaxis], 3, axis=2)
|
||||
imgray = cv2.cvtColor(cnts_images, cv2.COLOR_BGR2GRAY)
|
||||
ret, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
contours_imgs, hierarchy = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
|
||||
contours_imgs = return_parent_contours(contours_imgs, hierarchy)
|
||||
|
|
@ -243,123 +281,51 @@ def return_contours_of_interested_textline(region_pre_p, label):
|
|||
|
||||
def return_contours_of_image(image):
|
||||
if len(image.shape) == 2:
|
||||
image = np.repeat(image[:, :, np.newaxis], 3, axis=2)
|
||||
image = image.astype(np.uint8)
|
||||
imgray = image
|
||||
else:
|
||||
image = image.astype(np.uint8)
|
||||
imgray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
|
||||
_, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
imgray = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
|
||||
ret, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
contours, hierarchy = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
return contours, hierarchy
|
||||
|
||||
def dilate_textline_contours(all_found_textline_polygons):
|
||||
return [[polygon2contour(contour2polygon(contour, dilate=6))
|
||||
for contour in region]
|
||||
for region in all_found_textline_polygons]
|
||||
def return_contours_of_interested_region_by_min_size(region_pre_p, pixel, min_size=0.00003):
|
||||
# pixels of images are identified by 5
|
||||
if len(region_pre_p.shape) == 3:
|
||||
cnts_images = (region_pre_p[:, :, 0] == pixel) * 1
|
||||
else:
|
||||
cnts_images = (region_pre_p[:, :] == pixel) * 1
|
||||
cnts_images = cnts_images.astype(np.uint8)
|
||||
cnts_images = np.repeat(cnts_images[:, :, np.newaxis], 3, axis=2)
|
||||
imgray = cv2.cvtColor(cnts_images, cv2.COLOR_BGR2GRAY)
|
||||
ret, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
|
||||
def dilate_textregion_contours(all_found_textline_polygons):
|
||||
return [polygon2contour(contour2polygon(contour, dilate=6))
|
||||
for contour in all_found_textline_polygons]
|
||||
contours_imgs, hierarchy = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
contours_imgs = return_parent_contours(contours_imgs, hierarchy)
|
||||
contours_imgs = filter_contours_area_of_image_tables(
|
||||
thresh, contours_imgs, hierarchy, max_area=1, min_area=min_size)
|
||||
|
||||
def contour2polygon(contour: Union[np.ndarray, Sequence[Sequence[Sequence[Number]]]], dilate=0):
|
||||
polygon = Polygon([point[0] for point in contour])
|
||||
if dilate:
|
||||
polygon = polygon.buffer(dilate)
|
||||
if polygon.geom_type == 'GeometryCollection':
|
||||
# heterogeneous result: filter zero-area shapes (LineString, Point)
|
||||
polygon = unary_union([geom for geom in polygon.geoms if geom.area > 0])
|
||||
if polygon.geom_type == 'MultiPolygon':
|
||||
# homogeneous result: construct convex hull to connect
|
||||
polygon = join_polygons(polygon.geoms)
|
||||
return make_valid(polygon)
|
||||
return contours_imgs
|
||||
|
||||
def polygon2contour(polygon: Polygon) -> np.ndarray:
|
||||
polygon = np.array(polygon.exterior.coords[:-1], dtype=int)
|
||||
return np.maximum(0, polygon).astype(int)[:, np.newaxis]
|
||||
def return_contours_of_interested_region_by_size(region_pre_p, pixel, min_area, max_area):
|
||||
# pixels of images are identified by 5
|
||||
if len(region_pre_p.shape) == 3:
|
||||
cnts_images = (region_pre_p[:, :, 0] == pixel) * 1
|
||||
else:
|
||||
cnts_images = (region_pre_p[:, :] == pixel) * 1
|
||||
cnts_images = cnts_images.astype(np.uint8)
|
||||
cnts_images = np.repeat(cnts_images[:, :, np.newaxis], 3, axis=2)
|
||||
imgray = cv2.cvtColor(cnts_images, cv2.COLOR_BGR2GRAY)
|
||||
ret, thresh = cv2.threshold(imgray, 0, 255, 0)
|
||||
contours_imgs, hierarchy = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
|
||||
|
||||
def make_intersection(poly1, poly2):
|
||||
interp = poly1.intersection(poly2)
|
||||
# post-process
|
||||
if interp.is_empty or interp.area == 0.0:
|
||||
return None
|
||||
if interp.geom_type == 'GeometryCollection':
|
||||
# heterogeneous result: filter zero-area shapes (LineString, Point)
|
||||
interp = unary_union([geom for geom in interp.geoms if geom.area > 0])
|
||||
if interp.geom_type == 'MultiPolygon':
|
||||
# homogeneous result: construct convex hull to connect
|
||||
interp = join_polygons(interp.geoms)
|
||||
assert interp.geom_type == 'Polygon', interp.wkt
|
||||
interp = make_valid(interp)
|
||||
return interp
|
||||
contours_imgs = return_parent_contours(contours_imgs, hierarchy)
|
||||
contours_imgs = filter_contours_area_of_image_tables(
|
||||
thresh, contours_imgs, hierarchy, max_area=max_area, min_area=min_area)
|
||||
|
||||
def make_valid(polygon: Polygon) -> Polygon:
|
||||
"""Ensures shapely.geometry.Polygon object is valid by repeated rearrangement/simplification/enlargement."""
|
||||
def isint(x):
|
||||
return isinstance(x, int) or int(x) == x
|
||||
# make sure rounding does not invalidate
|
||||
if not all(map(isint, np.array(polygon.exterior.coords).flat)) and polygon.minimum_clearance < 1.0:
|
||||
polygon = Polygon(np.round(polygon.exterior.coords))
|
||||
points = list(polygon.exterior.coords[:-1])
|
||||
# try by re-arranging points
|
||||
for split in range(1, len(points)):
|
||||
if polygon.is_valid or polygon.simplify(polygon.area).is_valid:
|
||||
break
|
||||
# simplification may not be possible (at all) due to ordering
|
||||
# in that case, try another starting point
|
||||
polygon = Polygon(points[-split:]+points[:-split])
|
||||
# try by simplification
|
||||
for tolerance in range(int(polygon.area + 1.5)):
|
||||
if polygon.is_valid:
|
||||
break
|
||||
# simplification may require a larger tolerance
|
||||
polygon = polygon.simplify(tolerance + 1)
|
||||
# try by enlarging
|
||||
for tolerance in range(1, int(polygon.area + 2.5)):
|
||||
if polygon.is_valid:
|
||||
break
|
||||
# enlargement may require a larger tolerance
|
||||
polygon = polygon.buffer(tolerance)
|
||||
assert polygon.is_valid, polygon.wkt
|
||||
return polygon
|
||||
img_ret = np.zeros((region_pre_p.shape[0], region_pre_p.shape[1], 3))
|
||||
img_ret = cv2.fillPoly(img_ret, pts=contours_imgs, color=(1, 1, 1))
|
||||
|
||||
return img_ret[:, :, 0]
|
||||
|
||||
def join_polygons(polygons: Sequence[Polygon], scale=20) -> Polygon:
|
||||
"""construct concave hull (alpha shape) from input polygons by connecting their pairwise nearest points"""
|
||||
# ensure input polygons are simply typed and all oriented equally
|
||||
polygons = [orient(poly)
|
||||
for poly in itertools.chain.from_iterable(
|
||||
[poly.geoms
|
||||
if poly.geom_type in ['MultiPolygon', 'GeometryCollection']
|
||||
else [poly]
|
||||
for poly in polygons])]
|
||||
npoly = len(polygons)
|
||||
if npoly == 1:
|
||||
return polygons[0]
|
||||
# find min-dist path through all polygons (travelling salesman)
|
||||
pairs = itertools.combinations(range(npoly), 2)
|
||||
dists = np.zeros((npoly, npoly), dtype=float)
|
||||
for i, j in pairs:
|
||||
dist = polygons[i].distance(polygons[j])
|
||||
if dist < 1e-5:
|
||||
dist = 1e-5 # if pair merely touches, we still need to get an edge
|
||||
dists[i, j] = dist
|
||||
dists[j, i] = dist
|
||||
dists = minimum_spanning_tree(dists, overwrite=True)
|
||||
# add bridge polygons (where necessary)
|
||||
for prevp, nextp in zip(*dists.nonzero()):
|
||||
prevp = polygons[prevp]
|
||||
nextp = polygons[nextp]
|
||||
nearest = nearest_points(prevp, nextp)
|
||||
bridgep = orient(LineString(nearest).buffer(max(1, scale/5), resolution=1), -1)
|
||||
polygons.append(bridgep)
|
||||
jointp = unary_union(polygons)
|
||||
if jointp.geom_type == 'MultiPolygon':
|
||||
jointp = unary_union(jointp.geoms)
|
||||
assert jointp.geom_type == 'Polygon', jointp.wkt
|
||||
# follow-up calculations will necessarily be integer;
|
||||
# so anticipate rounding here and then ensure validity
|
||||
jointp2 = set_precision(jointp, 1.0, mode="keep_collapsed")
|
||||
if jointp2.geom_type != 'Polygon' or not jointp2.is_valid:
|
||||
jointp2 = Polygon(np.round(jointp.exterior.coords))
|
||||
jointp2 = make_valid(jointp2)
|
||||
assert jointp2.geom_type == 'Polygon', jointp2.wkt
|
||||
return jointp2
|
||||
|
|
|
|||
|
|
@ -3,7 +3,7 @@ from collections import Counter
|
|||
REGION_ID_TEMPLATE = 'region_%04d'
|
||||
LINE_ID_TEMPLATE = 'region_%04d_line_%04d'
|
||||
|
||||
class EynollahIdCounter:
|
||||
class EynollahIdCounter():
|
||||
|
||||
def __init__(self, region_idx=0, line_idx=0):
|
||||
self._counter = Counter()
|
||||
|
|
|
|||
|
|
@ -1,7 +1,6 @@
|
|||
import numpy as np
|
||||
import cv2
|
||||
from .contour import (
|
||||
find_center_of_contours,
|
||||
find_new_features_of_contours,
|
||||
return_contours_of_image,
|
||||
return_parent_contours,
|
||||
|
|
@ -19,11 +18,12 @@ def adhere_drop_capital_region_into_corresponding_textline(
|
|||
all_found_textline_polygons_h,
|
||||
kernel=None,
|
||||
curved_line=False,
|
||||
textline_light=False,
|
||||
):
|
||||
# print(np.shape(all_found_textline_polygons),np.shape(all_found_textline_polygons[3]),'all_found_textline_polygonsshape')
|
||||
# print(all_found_textline_polygons[3])
|
||||
cx_m, cy_m = find_center_of_contours(contours_only_text_parent)
|
||||
cx_h, cy_h = find_center_of_contours(contours_only_text_parent_h)
|
||||
cx_m, cy_m, _, _, _, _, _ = find_new_features_of_contours(contours_only_text_parent)
|
||||
cx_h, cy_h, _, _, _, _, _ = find_new_features_of_contours(contours_only_text_parent_h)
|
||||
cx_d, cy_d, _, _, y_min_d, y_max_d, _ = find_new_features_of_contours(polygons_of_drop_capitals)
|
||||
|
||||
img_con_all = np.zeros((text_regions_p.shape[0], text_regions_p.shape[1], 3))
|
||||
|
|
@ -78,7 +78,7 @@ def adhere_drop_capital_region_into_corresponding_textline(
|
|||
# region_with_intersected_drop=region_with_intersected_drop/3
|
||||
region_with_intersected_drop = region_with_intersected_drop.astype(np.uint8)
|
||||
# print(np.unique(img_con_all_copy[:,:,0]))
|
||||
if curved_line:
|
||||
if curved_line or textline_light:
|
||||
|
||||
if len(region_with_intersected_drop) > 1:
|
||||
sum_pixels_of_intersection = []
|
||||
|
|
@ -89,9 +89,9 @@ def adhere_drop_capital_region_into_corresponding_textline(
|
|||
region_final = region_with_intersected_drop[np.argmax(sum_pixels_of_intersection)] - 1
|
||||
|
||||
# print(region_final,'region_final')
|
||||
# cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
# cx_t,cy_t ,_, _, _ ,_,_= find_new_features_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
try:
|
||||
cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
cx_t, cy_t, _, _, _, _, _ = find_new_features_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
# print(all_box_coord[j_cont])
|
||||
# print(cx_t)
|
||||
# print(cy_t)
|
||||
|
|
@ -153,9 +153,9 @@ def adhere_drop_capital_region_into_corresponding_textline(
|
|||
|
||||
# areas_main=np.array([cv2.contourArea(all_found_textline_polygons[int(region_final)][0][j] ) for j in range(len(all_found_textline_polygons[int(region_final)]))])
|
||||
|
||||
# cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
# cx_t,cy_t ,_, _, _ ,_,_= find_new_features_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
try:
|
||||
cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
cx_t, cy_t, _, _, _, _, _ = find_new_features_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
# print(all_box_coord[j_cont])
|
||||
# print(cx_t)
|
||||
# print(cy_t)
|
||||
|
|
@ -208,7 +208,7 @@ def adhere_drop_capital_region_into_corresponding_textline(
|
|||
|
||||
try:
|
||||
# print(all_found_textline_polygons[j_cont][0])
|
||||
cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
cx_t, cy_t, _, _, _, _, _ = find_new_features_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
# print(all_box_coord[j_cont])
|
||||
# print(cx_t)
|
||||
# print(cy_t)
|
||||
|
|
@ -261,7 +261,7 @@ def adhere_drop_capital_region_into_corresponding_textline(
|
|||
else:
|
||||
pass
|
||||
|
||||
##cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
##cx_t,cy_t ,_, _, _ ,_,_= find_new_features_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
###print(all_box_coord[j_cont])
|
||||
###print(cx_t)
|
||||
###print(cy_t)
|
||||
|
|
@ -315,9 +315,9 @@ def adhere_drop_capital_region_into_corresponding_textline(
|
|||
region_final = region_with_intersected_drop[np.argmax(sum_pixels_of_intersection)] - 1
|
||||
|
||||
# print(region_final,'region_final')
|
||||
# cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
# cx_t,cy_t ,_, _, _ ,_,_= find_new_features_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
try:
|
||||
cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
cx_t, cy_t, _, _, _, _, _ = find_new_features_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
# print(all_box_coord[j_cont])
|
||||
# print(cx_t)
|
||||
# print(cy_t)
|
||||
|
|
@ -375,12 +375,12 @@ def adhere_drop_capital_region_into_corresponding_textline(
|
|||
|
||||
# areas_main=np.array([cv2.contourArea(all_found_textline_polygons[int(region_final)][0][j] ) for j in range(len(all_found_textline_polygons[int(region_final)]))])
|
||||
|
||||
# cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
# cx_t,cy_t ,_, _, _ ,_,_= find_new_features_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
|
||||
# print(cx_t,'print')
|
||||
try:
|
||||
# print(all_found_textline_polygons[j_cont][0])
|
||||
cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
cx_t, cy_t, _, _, _, _, _ = find_new_features_of_contours(all_found_textline_polygons[int(region_final)])
|
||||
# print(all_box_coord[j_cont])
|
||||
# print(cx_t)
|
||||
# print(cy_t)
|
||||
|
|
@ -453,7 +453,7 @@ def adhere_drop_capital_region_into_corresponding_textline(
|
|||
#####try:
|
||||
#####if len(contours_new_parent)==1:
|
||||
######print(all_found_textline_polygons[j_cont][0])
|
||||
#####cx_t, cy_t = find_center_of_contours(all_found_textline_polygons[j_cont])
|
||||
#####cx_t,cy_t ,_, _, _ ,_,_= find_new_features_of_contours(all_found_textline_polygons[j_cont])
|
||||
######print(all_box_coord[j_cont])
|
||||
######print(cx_t)
|
||||
######print(cy_t)
|
||||
|
|
|
|||
|
|
@ -1,16 +0,0 @@
|
|||
|
||||
# cannot use importlib.resources until we move to 3.9+ forimportlib.resources.files
|
||||
import sys
|
||||
from PIL import ImageFont
|
||||
|
||||
if sys.version_info < (3, 10):
|
||||
import importlib_resources
|
||||
else:
|
||||
import importlib.resources as importlib_resources
|
||||
|
||||
|
||||
def get_font():
|
||||
#font_path = "Charis-7.000/Charis-Regular.ttf" # Make sure this file exists!
|
||||
font = importlib_resources.files(__package__) / "../Charis-Regular.ttf"
|
||||
with importlib_resources.as_file(font) as font:
|
||||
return ImageFont.truetype(font=font, size=40)
|
||||
|
|
@ -6,10 +6,11 @@ from .contour import find_new_features_of_contours, return_contours_of_intereste
|
|||
from .resize import resize_image
|
||||
from .rotate import rotate_image
|
||||
|
||||
def get_marginals(text_with_lines, text_regions, num_col, slope_deskew, kernel=None):
|
||||
def get_marginals(text_with_lines, text_regions, num_col, slope_deskew, light_version=False, kernel=None):
|
||||
mask_marginals=np.zeros((text_with_lines.shape[0],text_with_lines.shape[1]))
|
||||
mask_marginals=mask_marginals.astype(np.uint8)
|
||||
|
||||
|
||||
text_with_lines=text_with_lines.astype(np.uint8)
|
||||
##text_with_lines=cv2.erode(text_with_lines,self.kernel,iterations=3)
|
||||
|
||||
|
|
@ -25,11 +26,8 @@ def get_marginals(text_with_lines, text_regions, num_col, slope_deskew, kernel=N
|
|||
text_with_lines=resize_image(text_with_lines,int(text_with_lines.shape[0]*1.8),text_with_lines.shape[1])
|
||||
text_with_lines=cv2.erode(text_with_lines,kernel,iterations=7)
|
||||
text_with_lines=resize_image(text_with_lines,text_with_lines_eroded.shape[0],text_with_lines_eroded.shape[1])
|
||||
|
||||
|
||||
kernel_hor = np.ones((1, 5), dtype=np.uint8)
|
||||
text_with_lines = cv2.erode(text_with_lines,kernel_hor,iterations=6)
|
||||
|
||||
|
||||
|
||||
text_with_lines_y=text_with_lines.sum(axis=0)
|
||||
text_with_lines_y_eroded=text_with_lines_eroded.sum(axis=0)
|
||||
|
||||
|
|
@ -42,7 +40,8 @@ def get_marginals(text_with_lines, text_regions, num_col, slope_deskew, kernel=N
|
|||
elif thickness_along_y_percent>=30 and thickness_along_y_percent<50:
|
||||
min_textline_thickness=20
|
||||
else:
|
||||
min_textline_thickness=45
|
||||
min_textline_thickness=40
|
||||
|
||||
|
||||
|
||||
if thickness_along_y_percent>=14:
|
||||
|
|
@ -72,7 +71,7 @@ def get_marginals(text_with_lines, text_regions, num_col, slope_deskew, kernel=N
|
|||
|
||||
peaks, _ = find_peaks(text_with_lines_y_rev, height=0)
|
||||
peaks=np.array(peaks)
|
||||
peaks=peaks[(peaks>first_nonzero) & (peaks < last_nonzero)]
|
||||
peaks=peaks[(peaks>first_nonzero) & ((peaks<last_nonzero))]
|
||||
peaks=peaks[region_sum_0[peaks]<min_textline_thickness ]
|
||||
|
||||
|
||||
|
|
@ -95,8 +94,6 @@ def get_marginals(text_with_lines, text_regions, num_col, slope_deskew, kernel=N
|
|||
except:
|
||||
point_left=first_nonzero
|
||||
|
||||
if point_left == first_nonzero and point_right == last_nonzero:
|
||||
return text_regions
|
||||
|
||||
|
||||
if point_right>=mask_marginals.shape[1]:
|
||||
|
|
@ -124,39 +121,92 @@ def get_marginals(text_with_lines, text_regions, num_col, slope_deskew, kernel=N
|
|||
if max_point_of_right_marginal>=text_regions.shape[1]:
|
||||
max_point_of_right_marginal=text_regions.shape[1]-1
|
||||
|
||||
text_regions_org = np.copy(text_regions)
|
||||
text_regions[text_regions[:,:]==1]=4
|
||||
|
||||
pixel_img=4
|
||||
min_area_text=0.00001
|
||||
|
||||
polygon_mask_marginals_rotated = return_contours_of_interested_region(mask_marginals,1,min_area_text)
|
||||
|
||||
polygon_mask_marginals_rotated = polygon_mask_marginals_rotated[0]
|
||||
if light_version:
|
||||
text_regions_org = np.copy(text_regions)
|
||||
text_regions[text_regions[:,:]==1]=4
|
||||
|
||||
pixel_img=4
|
||||
min_area_text=0.00001
|
||||
|
||||
polygon_mask_marginals_rotated = return_contours_of_interested_region(mask_marginals,1,min_area_text)
|
||||
|
||||
polygon_mask_marginals_rotated = polygon_mask_marginals_rotated[0]
|
||||
|
||||
polygons_of_marginals=return_contours_of_interested_region(text_regions,pixel_img,min_area_text)
|
||||
polygons_of_marginals=return_contours_of_interested_region(text_regions,pixel_img,min_area_text)
|
||||
|
||||
cx_text_only,cy_text_only ,x_min_text_only,x_max_text_only, y_min_text_only ,y_max_text_only,y_cor_x_min_main=find_new_features_of_contours(polygons_of_marginals)
|
||||
cx_text_only,cy_text_only ,x_min_text_only,x_max_text_only, y_min_text_only ,y_max_text_only,y_cor_x_min_main=find_new_features_of_contours(polygons_of_marginals)
|
||||
|
||||
text_regions[(text_regions[:,:]==4)]=1
|
||||
text_regions[(text_regions[:,:]==4)]=1
|
||||
|
||||
marginlas_should_be_main_text=[]
|
||||
marginlas_should_be_main_text=[]
|
||||
|
||||
x_min_marginals_left=[]
|
||||
x_min_marginals_right=[]
|
||||
x_min_marginals_left=[]
|
||||
x_min_marginals_right=[]
|
||||
|
||||
for i in range(len(cx_text_only)):
|
||||
results = cv2.pointPolygonTest(polygon_mask_marginals_rotated, (cx_text_only[i], cy_text_only[i]), False)
|
||||
for i in range(len(cx_text_only)):
|
||||
results = cv2.pointPolygonTest(polygon_mask_marginals_rotated, (cx_text_only[i], cy_text_only[i]), False)
|
||||
|
||||
if results == -1:
|
||||
marginlas_should_be_main_text.append(polygons_of_marginals[i])
|
||||
if results == -1:
|
||||
marginlas_should_be_main_text.append(polygons_of_marginals[i])
|
||||
|
||||
|
||||
|
||||
text_regions_org=cv2.fillPoly(text_regions_org, pts =marginlas_should_be_main_text, color=(4,4))
|
||||
text_regions = np.copy(text_regions_org)
|
||||
text_regions_org=cv2.fillPoly(text_regions_org, pts =marginlas_should_be_main_text, color=(4,4))
|
||||
text_regions = np.copy(text_regions_org)
|
||||
|
||||
|
||||
else:
|
||||
|
||||
text_regions[(mask_marginals_rotated[:,:]!=1) & (text_regions[:,:]==1)]=4
|
||||
|
||||
pixel_img=4
|
||||
min_area_text=0.00001
|
||||
|
||||
polygons_of_marginals=return_contours_of_interested_region(text_regions,pixel_img,min_area_text)
|
||||
|
||||
cx_text_only,cy_text_only ,x_min_text_only,x_max_text_only, y_min_text_only ,y_max_text_only,y_cor_x_min_main=find_new_features_of_contours(polygons_of_marginals)
|
||||
|
||||
text_regions[(text_regions[:,:]==4)]=1
|
||||
|
||||
marginlas_should_be_main_text=[]
|
||||
|
||||
x_min_marginals_left=[]
|
||||
x_min_marginals_right=[]
|
||||
|
||||
for i in range(len(cx_text_only)):
|
||||
x_width_mar=abs(x_min_text_only[i]-x_max_text_only[i])
|
||||
y_height_mar=abs(y_min_text_only[i]-y_max_text_only[i])
|
||||
|
||||
if x_width_mar>16 and y_height_mar/x_width_mar<18:
|
||||
marginlas_should_be_main_text.append(polygons_of_marginals[i])
|
||||
if x_min_text_only[i]<(mid_point-one_third_left):
|
||||
x_min_marginals_left_new=x_min_text_only[i]
|
||||
if len(x_min_marginals_left)==0:
|
||||
x_min_marginals_left.append(x_min_marginals_left_new)
|
||||
else:
|
||||
x_min_marginals_left[0]=min(x_min_marginals_left[0],x_min_marginals_left_new)
|
||||
else:
|
||||
x_min_marginals_right_new=x_min_text_only[i]
|
||||
if len(x_min_marginals_right)==0:
|
||||
x_min_marginals_right.append(x_min_marginals_right_new)
|
||||
else:
|
||||
x_min_marginals_right[0]=min(x_min_marginals_right[0],x_min_marginals_right_new)
|
||||
|
||||
if len(x_min_marginals_left)==0:
|
||||
x_min_marginals_left=[0]
|
||||
if len(x_min_marginals_right)==0:
|
||||
x_min_marginals_right=[text_regions.shape[1]-1]
|
||||
|
||||
|
||||
text_regions=cv2.fillPoly(text_regions, pts =marginlas_should_be_main_text, color=(4,4))
|
||||
|
||||
|
||||
#text_regions[:,:int(x_min_marginals_left[0])][text_regions[:,:int(x_min_marginals_left[0])]==1]=0
|
||||
#text_regions[:,int(x_min_marginals_right[0]):][text_regions[:,int(x_min_marginals_right[0]):]==1]=0
|
||||
|
||||
|
||||
text_regions[:,:int(min_point_of_left_marginal)][text_regions[:,:int(min_point_of_left_marginal)]==1]=0
|
||||
text_regions[:,int(max_point_of_right_marginal):][text_regions[:,int(max_point_of_right_marginal):]==1]=0
|
||||
|
||||
###text_regions[:,0:point_left][text_regions[:,0:point_left]==1]=4
|
||||
|
||||
|
|
|
|||
File diff suppressed because it is too large
Load diff
|
|
@ -1,45 +0,0 @@
|
|||
from multiprocessing import shared_memory
|
||||
from contextlib import contextmanager
|
||||
from functools import wraps
|
||||
import numpy as np
|
||||
|
||||
@contextmanager
|
||||
def share_ndarray(array: np.ndarray):
|
||||
size = np.dtype(array.dtype).itemsize * np.prod(array.shape)
|
||||
shm = shared_memory.SharedMemory(create=True, size=size)
|
||||
try:
|
||||
shared_array = np.ndarray(array.shape, dtype=array.dtype, buffer=shm.buf)
|
||||
shared_array[:] = array[:]
|
||||
shared_array.flags["WRITEABLE"] = False
|
||||
yield dict(shape=array.shape, dtype=array.dtype, name=shm.name)
|
||||
finally:
|
||||
shm.close()
|
||||
shm.unlink()
|
||||
|
||||
@contextmanager
|
||||
def ndarray_shared(array: dict):
|
||||
shm = shared_memory.SharedMemory(name=array['name'])
|
||||
try:
|
||||
array = np.ndarray(array['shape'], dtype=array['dtype'], buffer=shm.buf)
|
||||
yield array
|
||||
finally:
|
||||
shm.close()
|
||||
|
||||
def wrap_ndarray_shared(kw=None):
|
||||
def wrapper(f):
|
||||
if kw is None:
|
||||
@wraps(f)
|
||||
def shared_func(array, *args, **kwargs):
|
||||
with ndarray_shared(array) as ndarray:
|
||||
return f(ndarray, *args, **kwargs)
|
||||
return shared_func
|
||||
else:
|
||||
@wraps(f)
|
||||
def shared_func(*args, **kwargs):
|
||||
array = kwargs.pop(kw)
|
||||
with ndarray_shared(array) as ndarray:
|
||||
kwargs[kw] = ndarray
|
||||
return f(*args, **kwargs)
|
||||
return shared_func
|
||||
return wrapper
|
||||
|
||||
|
|
@ -1,510 +0,0 @@
|
|||
import math
|
||||
import copy
|
||||
|
||||
import numpy as np
|
||||
import cv2
|
||||
import tensorflow as tf
|
||||
from scipy.signal import find_peaks
|
||||
from scipy.ndimage import gaussian_filter1d
|
||||
from PIL import Image, ImageDraw, ImageFont
|
||||
from Bio import pairwise2
|
||||
|
||||
from .resize import resize_image
|
||||
|
||||
|
||||
def decode_batch_predictions(pred, num_to_char, max_len = 128):
|
||||
# input_len is the product of the batch size and the
|
||||
# number of time steps.
|
||||
input_len = np.ones(pred.shape[0]) * pred.shape[1]
|
||||
|
||||
# Decode CTC predictions using greedy search.
|
||||
# decoded is a tuple with 2 elements.
|
||||
decoded = tf.keras.backend.ctc_decode(pred,
|
||||
input_length = input_len,
|
||||
beam_width = 100)
|
||||
# The outputs are in the first element of the tuple.
|
||||
# Additionally, the first element is actually a list,
|
||||
# therefore we take the first element of that list as well.
|
||||
#print(decoded,'decoded')
|
||||
decoded = decoded[0][0][:, :max_len]
|
||||
|
||||
#print(decoded, decoded.shape,'decoded')
|
||||
|
||||
output = []
|
||||
for d in decoded:
|
||||
# Convert the predicted indices to the corresponding chars.
|
||||
d = tf.strings.reduce_join(num_to_char(d))
|
||||
d = d.numpy().decode("utf-8")
|
||||
output.append(d)
|
||||
return output
|
||||
|
||||
|
||||
def distortion_free_resize(image, img_size):
|
||||
w, h = img_size
|
||||
image = tf.image.resize(image, size=(h, w), preserve_aspect_ratio=True)
|
||||
|
||||
# Check tha amount of padding needed to be done.
|
||||
pad_height = h - tf.shape(image)[0]
|
||||
pad_width = w - tf.shape(image)[1]
|
||||
|
||||
# Only necessary if you want to do same amount of padding on both sides.
|
||||
if pad_height % 2 != 0:
|
||||
height = pad_height // 2
|
||||
pad_height_top = height + 1
|
||||
pad_height_bottom = height
|
||||
else:
|
||||
pad_height_top = pad_height_bottom = pad_height // 2
|
||||
|
||||
if pad_width % 2 != 0:
|
||||
width = pad_width // 2
|
||||
pad_width_left = width + 1
|
||||
pad_width_right = width
|
||||
else:
|
||||
pad_width_left = pad_width_right = pad_width // 2
|
||||
|
||||
image = tf.pad(
|
||||
image,
|
||||
paddings=[
|
||||
[pad_height_top, pad_height_bottom],
|
||||
[pad_width_left, pad_width_right],
|
||||
[0, 0],
|
||||
],
|
||||
)
|
||||
|
||||
image = tf.transpose(image, (1, 0, 2))
|
||||
image = tf.image.flip_left_right(image)
|
||||
return image
|
||||
|
||||
def return_start_and_end_of_common_text_of_textline_ocr_without_common_section(textline_image):
|
||||
width = np.shape(textline_image)[1]
|
||||
height = np.shape(textline_image)[0]
|
||||
common_window = int(0.06*width)
|
||||
|
||||
width1 = int ( width/2. - common_window )
|
||||
width2 = int ( width/2. + common_window )
|
||||
|
||||
img_sum = np.sum(textline_image[:,:,0], axis=0)
|
||||
sum_smoothed = gaussian_filter1d(img_sum, 3)
|
||||
|
||||
peaks_real, _ = find_peaks(sum_smoothed, height=0)
|
||||
if len(peaks_real)>70:
|
||||
|
||||
peaks_real = peaks_real[(peaks_real<width2) & (peaks_real>width1)]
|
||||
|
||||
arg_max = np.argmax(sum_smoothed[peaks_real])
|
||||
peaks_final = peaks_real[arg_max]
|
||||
return peaks_final
|
||||
else:
|
||||
return None
|
||||
|
||||
# Function to fit text inside the given area
|
||||
def fit_text_single_line(draw, text, font_path, max_width, max_height):
|
||||
initial_font_size = 50
|
||||
font_size = initial_font_size
|
||||
while font_size > 10: # Minimum font size
|
||||
font = ImageFont.truetype(font_path, font_size)
|
||||
text_bbox = draw.textbbox((0, 0), text, font=font) # Get text bounding box
|
||||
text_width = text_bbox[2] - text_bbox[0]
|
||||
text_height = text_bbox[3] - text_bbox[1]
|
||||
|
||||
if text_width <= max_width and text_height <= max_height:
|
||||
return font # Return the best-fitting font
|
||||
|
||||
font_size -= 2 # Reduce font size and retry
|
||||
|
||||
return ImageFont.truetype(font_path, 10) # Smallest font fallback
|
||||
|
||||
def return_textlines_split_if_needed(textline_image, textline_image_bin=None):
|
||||
|
||||
split_point = return_start_and_end_of_common_text_of_textline_ocr_without_common_section(textline_image)
|
||||
if split_point:
|
||||
image1 = textline_image[:, :split_point,:]# image.crop((0, 0, width2, height))
|
||||
image2 = textline_image[:, split_point:,:]#image.crop((width1, 0, width, height))
|
||||
if textline_image_bin is not None:
|
||||
image1_bin = textline_image_bin[:, :split_point,:]# image.crop((0, 0, width2, height))
|
||||
image2_bin = textline_image_bin[:, split_point:,:]#image.crop((width1, 0, width, height))
|
||||
return [image1, image2], [image1_bin, image2_bin]
|
||||
else:
|
||||
return [image1, image2], None
|
||||
else:
|
||||
return None, None
|
||||
|
||||
def preprocess_and_resize_image_for_ocrcnn_model(img, image_height, image_width):
|
||||
if img.shape[0]==0 or img.shape[1]==0:
|
||||
img_fin = np.ones((image_height, image_width, 3))
|
||||
else:
|
||||
ratio = image_height /float(img.shape[0])
|
||||
w_ratio = int(ratio * img.shape[1])
|
||||
|
||||
if w_ratio <= image_width:
|
||||
width_new = w_ratio
|
||||
else:
|
||||
width_new = image_width
|
||||
|
||||
if width_new == 0:
|
||||
width_new = img.shape[1]
|
||||
|
||||
|
||||
img = resize_image(img, image_height, width_new)
|
||||
img_fin = np.ones((image_height, image_width, 3))*255
|
||||
|
||||
img_fin[:,:width_new,:] = img[:,:,:]
|
||||
img_fin = img_fin / 255.
|
||||
return img_fin
|
||||
|
||||
def get_deskewed_contour_and_bb_and_image(contour, image, deskew_angle):
|
||||
(h_in, w_in) = image.shape[:2]
|
||||
center = (w_in // 2, h_in // 2)
|
||||
|
||||
rotation_matrix = cv2.getRotationMatrix2D(center, deskew_angle, 1.0)
|
||||
|
||||
cos_angle = abs(rotation_matrix[0, 0])
|
||||
sin_angle = abs(rotation_matrix[0, 1])
|
||||
new_w = int((h_in * sin_angle) + (w_in * cos_angle))
|
||||
new_h = int((h_in * cos_angle) + (w_in * sin_angle))
|
||||
|
||||
rotation_matrix[0, 2] += (new_w / 2) - center[0]
|
||||
rotation_matrix[1, 2] += (new_h / 2) - center[1]
|
||||
|
||||
deskewed_image = cv2.warpAffine(image, rotation_matrix, (new_w, new_h))
|
||||
|
||||
contour_points = np.array(contour, dtype=np.float32)
|
||||
transformed_points = cv2.transform(np.array([contour_points]), rotation_matrix)[0]
|
||||
|
||||
x, y, w, h = cv2.boundingRect(np.array(transformed_points, dtype=np.int32))
|
||||
cropped_textline = deskewed_image[y:y+h, x:x+w]
|
||||
|
||||
return cropped_textline
|
||||
|
||||
def rotate_image_with_padding(image, angle, border_value=(0,0,0)):
|
||||
# Get image dimensions
|
||||
(h, w) = image.shape[:2]
|
||||
|
||||
# Calculate the center of the image
|
||||
center = (w // 2, h // 2)
|
||||
|
||||
# Get the rotation matrix
|
||||
rotation_matrix = cv2.getRotationMatrix2D(center, angle, 1.0)
|
||||
|
||||
# Compute the new bounding dimensions
|
||||
cos = abs(rotation_matrix[0, 0])
|
||||
sin = abs(rotation_matrix[0, 1])
|
||||
new_w = int((h * sin) + (w * cos))
|
||||
new_h = int((h * cos) + (w * sin))
|
||||
|
||||
# Adjust the rotation matrix to account for translation
|
||||
rotation_matrix[0, 2] += (new_w / 2) - center[0]
|
||||
rotation_matrix[1, 2] += (new_h / 2) - center[1]
|
||||
|
||||
# Perform the rotation
|
||||
try:
|
||||
rotated_image = cv2.warpAffine(image, rotation_matrix, (new_w, new_h), borderValue=border_value)
|
||||
except:
|
||||
rotated_image = np.copy(image)
|
||||
|
||||
return rotated_image
|
||||
|
||||
def get_orientation_moments(contour):
|
||||
moments = cv2.moments(contour)
|
||||
if moments["mu20"] - moments["mu02"] == 0: # Avoid division by zero
|
||||
return 90 if moments["mu11"] > 0 else -90
|
||||
else:
|
||||
angle = 0.5 * np.arctan2(2 * moments["mu11"], moments["mu20"] - moments["mu02"])
|
||||
return np.degrees(angle) # Convert radians to degrees
|
||||
|
||||
|
||||
def get_orientation_moments_of_mask(mask):
|
||||
mask=mask.astype('uint8')
|
||||
contours, _ = cv2.findContours(mask[:,:,0], cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
|
||||
|
||||
largest_contour = max(contours, key=cv2.contourArea) if contours else None
|
||||
|
||||
moments = cv2.moments(largest_contour)
|
||||
if moments["mu20"] - moments["mu02"] == 0: # Avoid division by zero
|
||||
return 90 if moments["mu11"] > 0 else -90
|
||||
else:
|
||||
angle = 0.5 * np.arctan2(2 * moments["mu11"], moments["mu20"] - moments["mu02"])
|
||||
return np.degrees(angle) # Convert radians to degrees
|
||||
|
||||
def get_contours_and_bounding_boxes(mask):
|
||||
# Find contours in the binary mask
|
||||
contours, _ = cv2.findContours(mask, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
|
||||
|
||||
largest_contour = max(contours, key=cv2.contourArea) if contours else None
|
||||
|
||||
# Get the bounding rectangle for the contour
|
||||
x, y, w, h = cv2.boundingRect(largest_contour)
|
||||
#bounding_boxes.append((x, y, w, h))
|
||||
|
||||
return x, y, w, h
|
||||
|
||||
def return_splitting_point_of_image(image_to_spliited):
|
||||
width = np.shape(image_to_spliited)[1]
|
||||
height = np.shape(image_to_spliited)[0]
|
||||
common_window = int(0.03*width)
|
||||
|
||||
width1 = int ( common_window)
|
||||
width2 = int ( width - common_window )
|
||||
|
||||
img_sum = np.sum(image_to_spliited[:,:,0], axis=0)
|
||||
sum_smoothed = gaussian_filter1d(img_sum, 1)
|
||||
|
||||
peaks_real, _ = find_peaks(sum_smoothed, height=0)
|
||||
peaks_real = peaks_real[(peaks_real<width2) & (peaks_real>width1)]
|
||||
|
||||
arg_sort = np.argsort(sum_smoothed[peaks_real])
|
||||
peaks_sort_4 = peaks_real[arg_sort][::-1][:3]
|
||||
|
||||
return np.sort(peaks_sort_4)
|
||||
|
||||
def break_curved_line_into_small_pieces_and_then_merge(img_curved, mask_curved, img_bin_curved=None):
|
||||
peaks_4 = return_splitting_point_of_image(img_curved)
|
||||
if len(peaks_4)>0:
|
||||
imgs_tot = []
|
||||
|
||||
for ind in range(len(peaks_4)+1):
|
||||
if ind==0:
|
||||
img = img_curved[:, :peaks_4[ind], :]
|
||||
if img_bin_curved is not None:
|
||||
img_bin = img_bin_curved[:, :peaks_4[ind], :]
|
||||
mask = mask_curved[:, :peaks_4[ind], :]
|
||||
elif ind==len(peaks_4):
|
||||
img = img_curved[:, peaks_4[ind-1]:, :]
|
||||
if img_bin_curved is not None:
|
||||
img_bin = img_bin_curved[:, peaks_4[ind-1]:, :]
|
||||
mask = mask_curved[:, peaks_4[ind-1]:, :]
|
||||
else:
|
||||
img = img_curved[:, peaks_4[ind-1]:peaks_4[ind], :]
|
||||
if img_bin_curved is not None:
|
||||
img_bin = img_bin_curved[:, peaks_4[ind-1]:peaks_4[ind], :]
|
||||
mask = mask_curved[:, peaks_4[ind-1]:peaks_4[ind], :]
|
||||
|
||||
or_ma = get_orientation_moments_of_mask(mask)
|
||||
|
||||
if img_bin_curved is not None:
|
||||
imgs_tot.append([img, mask, or_ma, img_bin] )
|
||||
else:
|
||||
imgs_tot.append([img, mask, or_ma] )
|
||||
|
||||
|
||||
w_tot_des_list = []
|
||||
w_tot_des = 0
|
||||
imgs_deskewed_list = []
|
||||
imgs_bin_deskewed_list = []
|
||||
|
||||
for ind in range(len(imgs_tot)):
|
||||
img_in = imgs_tot[ind][0]
|
||||
mask_in = imgs_tot[ind][1]
|
||||
ori_in = imgs_tot[ind][2]
|
||||
if img_bin_curved is not None:
|
||||
img_bin_in = imgs_tot[ind][3]
|
||||
|
||||
if abs(ori_in)<45:
|
||||
img_in_des = rotate_image_with_padding(img_in, ori_in, border_value=(255,255,255) )
|
||||
if img_bin_curved is not None:
|
||||
img_bin_in_des = rotate_image_with_padding(img_bin_in, ori_in, border_value=(255,255,255) )
|
||||
mask_in_des = rotate_image_with_padding(mask_in, ori_in)
|
||||
mask_in_des = mask_in_des.astype('uint8')
|
||||
|
||||
#new bounding box
|
||||
x_n, y_n, w_n, h_n = get_contours_and_bounding_boxes(mask_in_des[:,:,0])
|
||||
|
||||
if w_n==0 or h_n==0:
|
||||
img_in_des = np.copy(img_in)
|
||||
if img_bin_curved is not None:
|
||||
img_bin_in_des = np.copy(img_bin_in)
|
||||
w_relative = int(32 * img_in_des.shape[1]/float(img_in_des.shape[0]) )
|
||||
if w_relative==0:
|
||||
w_relative = img_in_des.shape[1]
|
||||
img_in_des = resize_image(img_in_des, 32, w_relative)
|
||||
if img_bin_curved is not None:
|
||||
img_bin_in_des = resize_image(img_bin_in_des, 32, w_relative)
|
||||
else:
|
||||
mask_in_des = mask_in_des[y_n:y_n+h_n, x_n:x_n+w_n, :]
|
||||
img_in_des = img_in_des[y_n:y_n+h_n, x_n:x_n+w_n, :]
|
||||
if img_bin_curved is not None:
|
||||
img_bin_in_des = img_bin_in_des[y_n:y_n+h_n, x_n:x_n+w_n, :]
|
||||
|
||||
w_relative = int(32 * img_in_des.shape[1]/float(img_in_des.shape[0]) )
|
||||
if w_relative==0:
|
||||
w_relative = img_in_des.shape[1]
|
||||
img_in_des = resize_image(img_in_des, 32, w_relative)
|
||||
if img_bin_curved is not None:
|
||||
img_bin_in_des = resize_image(img_bin_in_des, 32, w_relative)
|
||||
|
||||
|
||||
else:
|
||||
img_in_des = np.copy(img_in)
|
||||
if img_bin_curved is not None:
|
||||
img_bin_in_des = np.copy(img_bin_in)
|
||||
w_relative = int(32 * img_in_des.shape[1]/float(img_in_des.shape[0]) )
|
||||
if w_relative==0:
|
||||
w_relative = img_in_des.shape[1]
|
||||
img_in_des = resize_image(img_in_des, 32, w_relative)
|
||||
if img_bin_curved is not None:
|
||||
img_bin_in_des = resize_image(img_bin_in_des, 32, w_relative)
|
||||
|
||||
w_tot_des+=img_in_des.shape[1]
|
||||
w_tot_des_list.append(img_in_des.shape[1])
|
||||
imgs_deskewed_list.append(img_in_des)
|
||||
if img_bin_curved is not None:
|
||||
imgs_bin_deskewed_list.append(img_bin_in_des)
|
||||
|
||||
|
||||
|
||||
|
||||
img_final_deskewed = np.zeros((32, w_tot_des, 3))+255
|
||||
if img_bin_curved is not None:
|
||||
img_bin_final_deskewed = np.zeros((32, w_tot_des, 3))+255
|
||||
else:
|
||||
img_bin_final_deskewed = None
|
||||
|
||||
w_indexer = 0
|
||||
for ind in range(len(w_tot_des_list)):
|
||||
img_final_deskewed[:,w_indexer:w_indexer+w_tot_des_list[ind],:] = imgs_deskewed_list[ind][:,:,:]
|
||||
if img_bin_curved is not None:
|
||||
img_bin_final_deskewed[:,w_indexer:w_indexer+w_tot_des_list[ind],:] = imgs_bin_deskewed_list[ind][:,:,:]
|
||||
w_indexer = w_indexer+w_tot_des_list[ind]
|
||||
return img_final_deskewed, img_bin_final_deskewed
|
||||
else:
|
||||
return img_curved, img_bin_curved
|
||||
|
||||
def return_textline_contour_with_added_box_coordinate(textline_contour, box_ind):
|
||||
textline_contour[:,0] = textline_contour[:,0] + box_ind[2]
|
||||
textline_contour[:,1] = textline_contour[:,1] + box_ind[0]
|
||||
return textline_contour
|
||||
|
||||
|
||||
def return_rnn_cnn_ocr_of_given_textlines(image,
|
||||
all_found_textline_polygons,
|
||||
all_box_coord,
|
||||
prediction_model,
|
||||
b_s_ocr, num_to_char,
|
||||
curved_line=False):
|
||||
max_len = 512
|
||||
padding_token = 299
|
||||
image_width = 512#max_len * 4
|
||||
image_height = 32
|
||||
ind_tot = 0
|
||||
#cv2.imwrite('./img_out.png', image_page)
|
||||
ocr_all_textlines = []
|
||||
cropped_lines_region_indexer = []
|
||||
cropped_lines_meging_indexing = []
|
||||
cropped_lines = []
|
||||
indexer_text_region = 0
|
||||
|
||||
for indexing, ind_poly_first in enumerate(all_found_textline_polygons):
|
||||
#ocr_textline_in_textregion = []
|
||||
if len(ind_poly_first)==0:
|
||||
cropped_lines_region_indexer.append(indexer_text_region)
|
||||
cropped_lines_meging_indexing.append(0)
|
||||
img_fin = np.ones((image_height, image_width, 3))*1
|
||||
cropped_lines.append(img_fin)
|
||||
|
||||
else:
|
||||
for indexing2, ind_poly in enumerate(ind_poly_first):
|
||||
cropped_lines_region_indexer.append(indexer_text_region)
|
||||
if not curved_line:
|
||||
ind_poly = copy.deepcopy(ind_poly)
|
||||
box_ind = all_box_coord[indexing]
|
||||
|
||||
ind_poly = return_textline_contour_with_added_box_coordinate(ind_poly, box_ind)
|
||||
#print(ind_poly_copy)
|
||||
ind_poly[ind_poly<0] = 0
|
||||
x, y, w, h = cv2.boundingRect(ind_poly)
|
||||
|
||||
w_scaled = w * image_height/float(h)
|
||||
|
||||
mask_poly = np.zeros(image.shape)
|
||||
|
||||
img_poly_on_img = np.copy(image)
|
||||
|
||||
mask_poly = cv2.fillPoly(mask_poly, pts=[ind_poly], color=(1, 1, 1))
|
||||
|
||||
|
||||
|
||||
mask_poly = mask_poly[y:y+h, x:x+w, :]
|
||||
img_crop = img_poly_on_img[y:y+h, x:x+w, :]
|
||||
|
||||
img_crop[mask_poly==0] = 255
|
||||
|
||||
if w_scaled < 640:#1.5*image_width:
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(img_crop, image_height, image_width)
|
||||
cropped_lines.append(img_fin)
|
||||
cropped_lines_meging_indexing.append(0)
|
||||
else:
|
||||
splited_images, splited_images_bin = return_textlines_split_if_needed(img_crop, None)
|
||||
|
||||
if splited_images:
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(splited_images[0],
|
||||
image_height,
|
||||
image_width)
|
||||
cropped_lines.append(img_fin)
|
||||
cropped_lines_meging_indexing.append(1)
|
||||
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(splited_images[1],
|
||||
image_height,
|
||||
image_width)
|
||||
|
||||
cropped_lines.append(img_fin)
|
||||
cropped_lines_meging_indexing.append(-1)
|
||||
|
||||
else:
|
||||
img_fin = preprocess_and_resize_image_for_ocrcnn_model(img_crop,
|
||||
image_height,
|
||||
image_width)
|
||||
cropped_lines.append(img_fin)
|
||||
cropped_lines_meging_indexing.append(0)
|
||||
|
||||
indexer_text_region+=1
|
||||
|
||||
extracted_texts = []
|
||||
|
||||
n_iterations = math.ceil(len(cropped_lines) / b_s_ocr)
|
||||
|
||||
for i in range(n_iterations):
|
||||
if i==(n_iterations-1):
|
||||
n_start = i*b_s_ocr
|
||||
imgs = cropped_lines[n_start:]
|
||||
imgs = np.array(imgs)
|
||||
imgs = imgs.reshape(imgs.shape[0], image_height, image_width, 3)
|
||||
|
||||
|
||||
else:
|
||||
n_start = i*b_s_ocr
|
||||
n_end = (i+1)*b_s_ocr
|
||||
imgs = cropped_lines[n_start:n_end]
|
||||
imgs = np.array(imgs).reshape(b_s_ocr, image_height, image_width, 3)
|
||||
|
||||
|
||||
preds = prediction_model.predict(imgs, verbose=0)
|
||||
|
||||
pred_texts = decode_batch_predictions(preds, num_to_char)
|
||||
|
||||
for ib in range(imgs.shape[0]):
|
||||
pred_texts_ib = pred_texts[ib].replace("[UNK]", "")
|
||||
extracted_texts.append(pred_texts_ib)
|
||||
|
||||
extracted_texts_merged = [extracted_texts[ind]
|
||||
if cropped_lines_meging_indexing[ind]==0
|
||||
else extracted_texts[ind]+" "+extracted_texts[ind+1]
|
||||
if cropped_lines_meging_indexing[ind]==1
|
||||
else None
|
||||
for ind in range(len(cropped_lines_meging_indexing))]
|
||||
|
||||
extracted_texts_merged = [ind for ind in extracted_texts_merged if ind is not None]
|
||||
unique_cropped_lines_region_indexer = np.unique(cropped_lines_region_indexer)
|
||||
|
||||
ocr_all_textlines = []
|
||||
for ind in unique_cropped_lines_region_indexer:
|
||||
ocr_textline_in_textregion = []
|
||||
extracted_texts_merged_un = np.array(extracted_texts_merged)[np.array(cropped_lines_region_indexer)==ind]
|
||||
for it_ind, text_textline in enumerate(extracted_texts_merged_un):
|
||||
ocr_textline_in_textregion.append(text_textline)
|
||||
ocr_all_textlines.append(ocr_textline_in_textregion)
|
||||
return ocr_all_textlines
|
||||
|
||||
def biopython_align(str1, str2):
|
||||
alignments = pairwise2.align.globalms(str1, str2, 2, -1, -2, -2)
|
||||
best_alignment = alignments[0] # Get the best alignment
|
||||
return best_alignment.seqA, best_alignment.seqB
|
||||
|
|
@ -46,26 +46,24 @@ def create_page_xml(imageFilename, height, width):
|
|||
))
|
||||
return pcgts
|
||||
|
||||
def xml_reading_order(page, order_of_texts, id_of_marginalia_left, id_of_marginalia_right):
|
||||
def xml_reading_order(page, order_of_texts, id_of_marginalia):
|
||||
region_order = ReadingOrderType()
|
||||
og = OrderedGroupType(id="ro357564684568544579089")
|
||||
page.set_ReadingOrder(region_order)
|
||||
region_order.set_OrderedGroup(og)
|
||||
region_counter = EynollahIdCounter()
|
||||
|
||||
for id_marginal in id_of_marginalia_left:
|
||||
og.add_RegionRefIndexed(RegionRefIndexedType(index=str(region_counter.get('region')), regionRef=id_marginal))
|
||||
for idx_textregion, _ in enumerate(order_of_texts):
|
||||
og.add_RegionRefIndexed(RegionRefIndexedType(index=str(region_counter.get('region')), regionRef=region_counter.region_id(order_of_texts[idx_textregion] + 1)))
|
||||
region_counter.inc('region')
|
||||
|
||||
for idx_textregion in order_of_texts:
|
||||
og.add_RegionRefIndexed(RegionRefIndexedType(index=str(region_counter.get('region')), regionRef=region_counter.region_id(idx_textregion + 1)))
|
||||
region_counter.inc('region')
|
||||
|
||||
for id_marginal in id_of_marginalia_right:
|
||||
for id_marginal in id_of_marginalia:
|
||||
og.add_RegionRefIndexed(RegionRefIndexedType(index=str(region_counter.get('region')), regionRef=id_marginal))
|
||||
region_counter.inc('region')
|
||||
|
||||
def order_and_id_of_texts(found_polygons_text_region, found_polygons_text_region_h, indexes_sorted, index_of_types, kind_of_texts, ref_point):
|
||||
def order_and_id_of_texts(found_polygons_text_region, found_polygons_text_region_h, matrix_of_orders, indexes_sorted, index_of_types, kind_of_texts, ref_point):
|
||||
indexes_sorted = np.array(indexes_sorted)
|
||||
index_of_types = np.array(index_of_types)
|
||||
kind_of_texts = np.array(kind_of_texts)
|
||||
|
||||
id_of_texts = []
|
||||
order_of_texts = []
|
||||
|
||||
|
|
@ -88,7 +86,3 @@ def order_and_id_of_texts(found_polygons_text_region, found_polygons_text_region
|
|||
order_of_texts.append(interest)
|
||||
|
||||
return order_of_texts, id_of_texts
|
||||
|
||||
def etree_namespace_for_element_tag(tag: str):
|
||||
right = tag.find('}')
|
||||
return tag[1:right]
|
||||
|
|
|
|||
|
|
@ -2,14 +2,15 @@
|
|||
# pylint: disable=import-error
|
||||
from pathlib import Path
|
||||
import os.path
|
||||
from typing import Optional
|
||||
import logging
|
||||
import xml.etree.ElementTree as ET
|
||||
from .utils.xml import create_page_xml, xml_reading_order
|
||||
from .utils.counter import EynollahIdCounter
|
||||
|
||||
from ocrd_utils import getLogger
|
||||
from ocrd_models.ocrd_page import (
|
||||
BorderType,
|
||||
CoordsType,
|
||||
PcGtsType,
|
||||
TextLineType,
|
||||
TextEquivType,
|
||||
TextRegionType,
|
||||
|
|
@ -18,21 +19,23 @@ from ocrd_models.ocrd_page import (
|
|||
SeparatorRegionType,
|
||||
to_xml
|
||||
)
|
||||
import numpy as np
|
||||
|
||||
class EynollahXmlWriter:
|
||||
class EynollahXmlWriter():
|
||||
|
||||
def __init__(self, *, dir_out, image_filename, curved_line, pcgts=None):
|
||||
self.logger = logging.getLogger('eynollah.writer')
|
||||
def __init__(self, *, dir_out, image_filename, curved_line,textline_light, pcgts=None):
|
||||
self.logger = getLogger('eynollah.writer')
|
||||
self.counter = EynollahIdCounter()
|
||||
self.dir_out = dir_out
|
||||
self.image_filename = image_filename
|
||||
self.output_filename = os.path.join(self.dir_out or "", self.image_filename_stem) + ".xml"
|
||||
self.curved_line = curved_line
|
||||
self.textline_light = textline_light
|
||||
self.pcgts = pcgts
|
||||
self.scale_x: Optional[float] = None # XXX set outside __init__
|
||||
self.scale_y: Optional[float] = None # XXX set outside __init__
|
||||
self.height_org: Optional[int] = None # XXX set outside __init__
|
||||
self.width_org: Optional[int] = None # XXX set outside __init__
|
||||
self.scale_x = None # XXX set outside __init__
|
||||
self.scale_y = None # XXX set outside __init__
|
||||
self.height_org = None # XXX set outside __init__
|
||||
self.width_org = None # XXX set outside __init__
|
||||
|
||||
@property
|
||||
def image_filename_stem(self):
|
||||
|
|
@ -53,26 +56,111 @@ class EynollahXmlWriter:
|
|||
points_page_print = points_page_print + ' '
|
||||
return points_page_print[:-1]
|
||||
|
||||
def serialize_lines_in_marginal(self, marginal_region, all_found_textline_polygons_marginals, marginal_idx, page_coord, all_box_coord_marginals, slopes_marginals, counter):
|
||||
for j in range(len(all_found_textline_polygons_marginals[marginal_idx])):
|
||||
coords = CoordsType()
|
||||
textline = TextLineType(id=counter.next_line_id, Coords=coords)
|
||||
marginal_region.add_TextLine(textline)
|
||||
marginal_region.set_orientation(-slopes_marginals[marginal_idx])
|
||||
points_co = ''
|
||||
for l in range(len(all_found_textline_polygons_marginals[marginal_idx][j])):
|
||||
if not (self.curved_line or self.textline_light):
|
||||
if len(all_found_textline_polygons_marginals[marginal_idx][j][l]) == 2:
|
||||
textline_x_coord = max(0, int((all_found_textline_polygons_marginals[marginal_idx][j][l][0] + all_box_coord_marginals[marginal_idx][2] + page_coord[2]) / self.scale_x) )
|
||||
textline_y_coord = max(0, int((all_found_textline_polygons_marginals[marginal_idx][j][l][1] + all_box_coord_marginals[marginal_idx][0] + page_coord[0]) / self.scale_y) )
|
||||
else:
|
||||
textline_x_coord = max(0, int((all_found_textline_polygons_marginals[marginal_idx][j][l][0][0] + all_box_coord_marginals[marginal_idx][2] + page_coord[2]) / self.scale_x) )
|
||||
textline_y_coord = max(0, int((all_found_textline_polygons_marginals[marginal_idx][j][l][0][1] + all_box_coord_marginals[marginal_idx][0] + page_coord[0]) / self.scale_y) )
|
||||
points_co += str(textline_x_coord)
|
||||
points_co += ','
|
||||
points_co += str(textline_y_coord)
|
||||
if (self.curved_line or self.textline_light) and np.abs(slopes_marginals[marginal_idx]) <= 45:
|
||||
if len(all_found_textline_polygons_marginals[marginal_idx][j][l]) == 2:
|
||||
points_co += str(int((all_found_textline_polygons_marginals[marginal_idx][j][l][0] + page_coord[2]) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((all_found_textline_polygons_marginals[marginal_idx][j][l][1] + page_coord[0]) / self.scale_y))
|
||||
else:
|
||||
points_co += str(int((all_found_textline_polygons_marginals[marginal_idx][j][l][0][0] + page_coord[2]) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((all_found_textline_polygons_marginals[marginal_idx][j][l][0][1] + page_coord[0]) / self.scale_y))
|
||||
|
||||
elif (self.curved_line or self.textline_light) and np.abs(slopes_marginals[marginal_idx]) > 45:
|
||||
if len(all_found_textline_polygons_marginals[marginal_idx][j][l]) == 2:
|
||||
points_co += str(int((all_found_textline_polygons_marginals[marginal_idx][j][l][0] + all_box_coord_marginals[marginal_idx][2] + page_coord[2]) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((all_found_textline_polygons_marginals[marginal_idx][j][l][1] + all_box_coord_marginals[marginal_idx][0] + page_coord[0]) / self.scale_y))
|
||||
else:
|
||||
points_co += str(int((all_found_textline_polygons_marginals[marginal_idx][j][l][0][0] + all_box_coord_marginals[marginal_idx][2] + page_coord[2]) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((all_found_textline_polygons_marginals[marginal_idx][j][l][0][1] + all_box_coord_marginals[marginal_idx][0] + page_coord[0]) / self.scale_y))
|
||||
points_co += ' '
|
||||
coords.set_points(points_co[:-1])
|
||||
|
||||
def serialize_lines_in_region(self, text_region, all_found_textline_polygons, region_idx, page_coord, all_box_coord, slopes, counter, ocr_all_textlines_textregion):
|
||||
self.logger.debug('enter serialize_lines_in_region')
|
||||
for j, polygon_textline in enumerate(all_found_textline_polygons[region_idx]):
|
||||
for j in range(len(all_found_textline_polygons[region_idx])):
|
||||
coords = CoordsType()
|
||||
textline = TextLineType(id=counter.next_line_id, Coords=coords)
|
||||
if ocr_all_textlines_textregion:
|
||||
# FIXME: add OCR confidence
|
||||
textline.set_TextEquiv([TextEquivType(Unicode=ocr_all_textlines_textregion[j])])
|
||||
textline.set_TextEquiv( [ TextEquivType(Unicode=ocr_all_textlines_textregion[j]) ] )
|
||||
text_region.add_TextLine(textline)
|
||||
text_region.set_orientation(-slopes[region_idx])
|
||||
region_bboxes = all_box_coord[region_idx]
|
||||
points_co = ''
|
||||
for point in polygon_textline:
|
||||
if len(point) != 2:
|
||||
point = point[0]
|
||||
point_x = point[0] + page_coord[2]
|
||||
point_y = point[1] + page_coord[0]
|
||||
point_x = max(0, int(point_x / self.scale_x))
|
||||
point_y = max(0, int(point_y / self.scale_y))
|
||||
points_co += f'{point_x},{point_y} '
|
||||
for idx_contour_textline, contour_textline in enumerate(all_found_textline_polygons[region_idx][j]):
|
||||
if not (self.curved_line or self.textline_light):
|
||||
if len(contour_textline) == 2:
|
||||
textline_x_coord = max(0, int((contour_textline[0] + region_bboxes[2] + page_coord[2]) / self.scale_x))
|
||||
textline_y_coord = max(0, int((contour_textline[1] + region_bboxes[0] + page_coord[0]) / self.scale_y))
|
||||
else:
|
||||
textline_x_coord = max(0, int((contour_textline[0][0] + region_bboxes[2] + page_coord[2]) / self.scale_x))
|
||||
textline_y_coord = max(0, int((contour_textline[0][1] + region_bboxes[0] + page_coord[0]) / self.scale_y))
|
||||
points_co += str(textline_x_coord)
|
||||
points_co += ','
|
||||
points_co += str(textline_y_coord)
|
||||
|
||||
if (self.curved_line or self.textline_light) and np.abs(slopes[region_idx]) <= 45:
|
||||
if len(contour_textline) == 2:
|
||||
points_co += str(int((contour_textline[0] + page_coord[2]) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((contour_textline[1] + page_coord[0]) / self.scale_y))
|
||||
else:
|
||||
points_co += str(int((contour_textline[0][0] + page_coord[2]) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((contour_textline[0][1] + page_coord[0])/self.scale_y))
|
||||
elif (self.curved_line or self.textline_light) and np.abs(slopes[region_idx]) > 45:
|
||||
if len(contour_textline)==2:
|
||||
points_co += str(int((contour_textline[0] + region_bboxes[2] + page_coord[2])/self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((contour_textline[1] + region_bboxes[0] + page_coord[0])/self.scale_y))
|
||||
else:
|
||||
points_co += str(int((contour_textline[0][0] + region_bboxes[2]+page_coord[2])/self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((contour_textline[0][1] + region_bboxes[0]+page_coord[0])/self.scale_y))
|
||||
points_co += ' '
|
||||
coords.set_points(points_co[:-1])
|
||||
|
||||
def serialize_lines_in_dropcapital(self, text_region, all_found_textline_polygons, region_idx, page_coord, all_box_coord, slopes, counter, ocr_all_textlines_textregion):
|
||||
self.logger.debug('enter serialize_lines_in_region')
|
||||
for j in range(1):
|
||||
coords = CoordsType()
|
||||
textline = TextLineType(id=counter.next_line_id, Coords=coords)
|
||||
if ocr_all_textlines_textregion:
|
||||
textline.set_TextEquiv( [ TextEquivType(Unicode=ocr_all_textlines_textregion[j]) ] )
|
||||
text_region.add_TextLine(textline)
|
||||
#region_bboxes = all_box_coord[region_idx]
|
||||
points_co = ''
|
||||
for idx_contour_textline, contour_textline in enumerate(all_found_textline_polygons[j]):
|
||||
if len(contour_textline) == 2:
|
||||
points_co += str(int((contour_textline[0] + page_coord[2]) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((contour_textline[1] + page_coord[0]) / self.scale_y))
|
||||
else:
|
||||
points_co += str(int((contour_textline[0][0] + page_coord[2]) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((contour_textline[0][1] + page_coord[0])/self.scale_y))
|
||||
|
||||
points_co += ' '
|
||||
coords.set_points(points_co[:-1])
|
||||
|
||||
def write_pagexml(self, pcgts):
|
||||
|
|
@ -80,210 +168,153 @@ class EynollahXmlWriter:
|
|||
with open(self.output_filename, 'w') as f:
|
||||
f.write(to_xml(pcgts))
|
||||
|
||||
def build_pagexml_no_full_layout(
|
||||
self,
|
||||
*,
|
||||
found_polygons_text_region,
|
||||
page_coord,
|
||||
order_of_texts,
|
||||
all_found_textline_polygons,
|
||||
all_box_coord,
|
||||
found_polygons_text_region_img,
|
||||
found_polygons_marginals_left,
|
||||
found_polygons_marginals_right,
|
||||
all_found_textline_polygons_marginals_left,
|
||||
all_found_textline_polygons_marginals_right,
|
||||
all_box_coord_marginals_left,
|
||||
all_box_coord_marginals_right,
|
||||
slopes,
|
||||
slopes_marginals_left,
|
||||
slopes_marginals_right,
|
||||
cont_page,
|
||||
polygons_seplines,
|
||||
found_polygons_tables,
|
||||
):
|
||||
return self.build_pagexml_full_layout(
|
||||
found_polygons_text_region=found_polygons_text_region,
|
||||
found_polygons_text_region_h=[],
|
||||
page_coord=page_coord,
|
||||
order_of_texts=order_of_texts,
|
||||
all_found_textline_polygons=all_found_textline_polygons,
|
||||
all_found_textline_polygons_h=[],
|
||||
all_box_coord=all_box_coord,
|
||||
all_box_coord_h=[],
|
||||
found_polygons_text_region_img=found_polygons_text_region_img,
|
||||
found_polygons_tables=found_polygons_tables,
|
||||
found_polygons_drop_capitals=[],
|
||||
found_polygons_marginals_left=found_polygons_marginals_left,
|
||||
found_polygons_marginals_right=found_polygons_marginals_right,
|
||||
all_found_textline_polygons_marginals_left=all_found_textline_polygons_marginals_left,
|
||||
all_found_textline_polygons_marginals_right=all_found_textline_polygons_marginals_right,
|
||||
all_box_coord_marginals_left=all_box_coord_marginals_left,
|
||||
all_box_coord_marginals_right=all_box_coord_marginals_right,
|
||||
slopes=slopes,
|
||||
slopes_h=[],
|
||||
slopes_marginals_left=slopes_marginals_left,
|
||||
slopes_marginals_right=slopes_marginals_right,
|
||||
cont_page=cont_page,
|
||||
polygons_seplines=polygons_seplines,
|
||||
)
|
||||
|
||||
def build_pagexml_full_layout(
|
||||
self,
|
||||
*,
|
||||
found_polygons_text_region,
|
||||
found_polygons_text_region_h,
|
||||
page_coord,
|
||||
order_of_texts,
|
||||
all_found_textline_polygons,
|
||||
all_found_textline_polygons_h,
|
||||
all_box_coord,
|
||||
all_box_coord_h,
|
||||
found_polygons_text_region_img,
|
||||
found_polygons_tables,
|
||||
found_polygons_drop_capitals,
|
||||
found_polygons_marginals_left,
|
||||
found_polygons_marginals_right,
|
||||
all_found_textline_polygons_marginals_left,
|
||||
all_found_textline_polygons_marginals_right,
|
||||
all_box_coord_marginals_left,
|
||||
all_box_coord_marginals_right,
|
||||
slopes,
|
||||
slopes_h,
|
||||
slopes_marginals_left,
|
||||
slopes_marginals_right,
|
||||
cont_page,
|
||||
polygons_seplines,
|
||||
ocr_all_textlines=None,
|
||||
ocr_all_textlines_h=None,
|
||||
ocr_all_textlines_marginals_left=None,
|
||||
ocr_all_textlines_marginals_right=None,
|
||||
ocr_all_textlines_drop=None,
|
||||
conf_contours_textregions=None,
|
||||
conf_contours_textregions_h=None,
|
||||
skip_layout_reading_order=False,
|
||||
):
|
||||
self.logger.debug('enter build_pagexml')
|
||||
def build_pagexml_no_full_layout(self, found_polygons_text_region, page_coord, order_of_texts, id_of_texts, all_found_textline_polygons, all_box_coord, found_polygons_text_region_img, found_polygons_marginals, all_found_textline_polygons_marginals, all_box_coord_marginals, slopes, slopes_marginals, cont_page, polygons_lines_to_be_written_in_xml, found_polygons_tables, ocr_all_textlines, conf_contours_textregion):
|
||||
self.logger.debug('enter build_pagexml_no_full_layout')
|
||||
|
||||
# create the file structure
|
||||
pcgts = self.pcgts if self.pcgts else create_page_xml(self.image_filename, self.height_org, self.width_org)
|
||||
page = pcgts.get_Page()
|
||||
assert page
|
||||
page.set_Border(BorderType(Coords=CoordsType(points=self.calculate_page_coords(cont_page))))
|
||||
|
||||
counter = EynollahIdCounter()
|
||||
if len(order_of_texts):
|
||||
if len(found_polygons_text_region) > 0:
|
||||
_counter_marginals = EynollahIdCounter(region_idx=len(order_of_texts))
|
||||
id_of_marginalia_left = [_counter_marginals.next_region_id
|
||||
for _ in found_polygons_marginals_left]
|
||||
id_of_marginalia_right = [_counter_marginals.next_region_id
|
||||
for _ in found_polygons_marginals_right]
|
||||
xml_reading_order(page, order_of_texts, id_of_marginalia_left, id_of_marginalia_right)
|
||||
id_of_marginalia = [_counter_marginals.next_region_id for _ in found_polygons_marginals]
|
||||
xml_reading_order(page, order_of_texts, id_of_marginalia)
|
||||
|
||||
for mm, region_contour in enumerate(found_polygons_text_region):
|
||||
textregion = TextRegionType(
|
||||
id=counter.next_region_id, type_='paragraph',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(region_contour, page_coord,
|
||||
skip_layout_reading_order))
|
||||
)
|
||||
assert textregion.Coords
|
||||
if conf_contours_textregions:
|
||||
textregion.Coords.set_conf(conf_contours_textregions[mm])
|
||||
for mm in range(len(found_polygons_text_region)):
|
||||
textregion = TextRegionType(id=counter.next_region_id, type_='paragraph',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(found_polygons_text_region[mm], page_coord), conf=conf_contours_textregion[mm]),
|
||||
)
|
||||
#textregion.set_conf(conf_contours_textregion[mm])
|
||||
page.add_TextRegion(textregion)
|
||||
if ocr_all_textlines:
|
||||
ocr_textlines = ocr_all_textlines[mm]
|
||||
else:
|
||||
ocr_textlines = None
|
||||
self.serialize_lines_in_region(textregion, all_found_textline_polygons, mm, page_coord,
|
||||
all_box_coord, slopes, counter, ocr_textlines)
|
||||
self.serialize_lines_in_region(textregion, all_found_textline_polygons, mm, page_coord, all_box_coord, slopes, counter, ocr_textlines)
|
||||
|
||||
self.logger.debug('len(found_polygons_text_region_h) %s', len(found_polygons_text_region_h))
|
||||
for mm, region_contour in enumerate(found_polygons_text_region_h):
|
||||
textregion = TextRegionType(
|
||||
id=counter.next_region_id, type_='heading',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(region_contour, page_coord))
|
||||
)
|
||||
assert textregion.Coords
|
||||
if conf_contours_textregions_h:
|
||||
textregion.Coords.set_conf(conf_contours_textregions_h[mm])
|
||||
page.add_TextRegion(textregion)
|
||||
if ocr_all_textlines_h:
|
||||
ocr_textlines = ocr_all_textlines_h[mm]
|
||||
else:
|
||||
ocr_textlines = None
|
||||
self.serialize_lines_in_region(textregion, all_found_textline_polygons_h, mm, page_coord,
|
||||
all_box_coord_h, slopes_h, counter, ocr_textlines)
|
||||
|
||||
for mm, region_contour in enumerate(found_polygons_marginals_left):
|
||||
marginal = TextRegionType(
|
||||
id=counter.next_region_id, type_='marginalia',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(region_contour, page_coord))
|
||||
)
|
||||
for mm in range(len(found_polygons_marginals)):
|
||||
marginal = TextRegionType(id=counter.next_region_id, type_='marginalia',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(found_polygons_marginals[mm], page_coord)))
|
||||
page.add_TextRegion(marginal)
|
||||
if ocr_all_textlines_marginals_left:
|
||||
ocr_textlines = ocr_all_textlines_marginals_left[mm]
|
||||
else:
|
||||
ocr_textlines = None
|
||||
self.serialize_lines_in_region(marginal, all_found_textline_polygons_marginals_left, mm, page_coord, all_box_coord_marginals_left, slopes_marginals_left, counter, ocr_textlines)
|
||||
self.serialize_lines_in_marginal(marginal, all_found_textline_polygons_marginals, mm, page_coord, all_box_coord_marginals, slopes_marginals, counter)
|
||||
|
||||
for mm, region_contour in enumerate(found_polygons_marginals_right):
|
||||
marginal = TextRegionType(
|
||||
id=counter.next_region_id, type_='marginalia',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(region_contour, page_coord))
|
||||
)
|
||||
page.add_TextRegion(marginal)
|
||||
if ocr_all_textlines_marginals_right:
|
||||
ocr_textlines = ocr_all_textlines_marginals_right[mm]
|
||||
else:
|
||||
ocr_textlines = None
|
||||
self.serialize_lines_in_region(marginal, all_found_textline_polygons_marginals_right, mm, page_coord,
|
||||
all_box_coord_marginals_right, slopes_marginals_right, counter, ocr_textlines)
|
||||
for mm in range(len(found_polygons_text_region_img)):
|
||||
img_region = ImageRegionType(id=counter.next_region_id, Coords=CoordsType())
|
||||
page.add_ImageRegion(img_region)
|
||||
points_co = ''
|
||||
for lmm in range(len(found_polygons_text_region_img[mm])):
|
||||
try:
|
||||
points_co += str(int((found_polygons_text_region_img[mm][lmm,0,0] + page_coord[2]) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((found_polygons_text_region_img[mm][lmm,0,1] + page_coord[0]) / self.scale_y))
|
||||
points_co += ' '
|
||||
except:
|
||||
|
||||
for mm, region_contour in enumerate(found_polygons_drop_capitals):
|
||||
dropcapital = TextRegionType(
|
||||
id=counter.next_region_id, type_='drop-capital',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(region_contour, page_coord))
|
||||
)
|
||||
page.add_TextRegion(dropcapital)
|
||||
all_box_coord_drop = [[0, 0, 0, 0]]
|
||||
slopes_drop = [0]
|
||||
if ocr_all_textlines_drop:
|
||||
ocr_textlines = ocr_all_textlines_drop[mm]
|
||||
else:
|
||||
ocr_textlines = None
|
||||
self.serialize_lines_in_region(dropcapital, [[found_polygons_drop_capitals[mm]]], 0, page_coord,
|
||||
all_box_coord_drop, slopes_drop, counter, ocr_textlines)
|
||||
points_co += str(int((found_polygons_text_region_img[mm][lmm][0] + page_coord[2])/ self.scale_x ))
|
||||
points_co += ','
|
||||
points_co += str(int((found_polygons_text_region_img[mm][lmm][1] + page_coord[0])/ self.scale_y ))
|
||||
points_co += ' '
|
||||
|
||||
for region_contour in found_polygons_text_region_img:
|
||||
page.add_ImageRegion(
|
||||
ImageRegionType(id=counter.next_region_id,
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(region_contour, page_coord))))
|
||||
img_region.get_Coords().set_points(points_co[:-1])
|
||||
|
||||
for region_contour in polygons_seplines:
|
||||
page.add_SeparatorRegion(
|
||||
SeparatorRegionType(id=counter.next_region_id,
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(region_contour, [0, 0, 0, 0]))))
|
||||
|
||||
for region_contour in found_polygons_tables:
|
||||
page.add_TableRegion(
|
||||
TableRegionType(id=counter.next_region_id,
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(region_contour, page_coord))))
|
||||
for mm in range(len(polygons_lines_to_be_written_in_xml)):
|
||||
sep_hor = SeparatorRegionType(id=counter.next_region_id, Coords=CoordsType())
|
||||
page.add_SeparatorRegion(sep_hor)
|
||||
points_co = ''
|
||||
for lmm in range(len(polygons_lines_to_be_written_in_xml[mm])):
|
||||
points_co += str(int((polygons_lines_to_be_written_in_xml[mm][lmm,0,0] ) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((polygons_lines_to_be_written_in_xml[mm][lmm,0,1] ) / self.scale_y))
|
||||
points_co += ' '
|
||||
sep_hor.get_Coords().set_points(points_co[:-1])
|
||||
for mm in range(len(found_polygons_tables)):
|
||||
tab_region = TableRegionType(id=counter.next_region_id, Coords=CoordsType())
|
||||
page.add_TableRegion(tab_region)
|
||||
points_co = ''
|
||||
for lmm in range(len(found_polygons_tables[mm])):
|
||||
points_co += str(int((found_polygons_tables[mm][lmm,0,0] + page_coord[2]) / self.scale_x))
|
||||
points_co += ','
|
||||
points_co += str(int((found_polygons_tables[mm][lmm,0,1] + page_coord[0]) / self.scale_y))
|
||||
points_co += ' '
|
||||
tab_region.get_Coords().set_points(points_co[:-1])
|
||||
|
||||
return pcgts
|
||||
|
||||
def calculate_polygon_coords(self, contour, page_coord, skip_layout_reading_order=False):
|
||||
def build_pagexml_full_layout(self, found_polygons_text_region, found_polygons_text_region_h, page_coord, order_of_texts, id_of_texts, all_found_textline_polygons, all_found_textline_polygons_h, all_box_coord, all_box_coord_h, found_polygons_text_region_img, found_polygons_tables, found_polygons_drop_capitals, found_polygons_marginals, all_found_textline_polygons_marginals, all_box_coord_marginals, slopes, slopes_h, slopes_marginals, cont_page, polygons_lines_to_be_written_in_xml, ocr_all_textlines, conf_contours_textregion, conf_contours_textregion_h):
|
||||
self.logger.debug('enter build_pagexml_full_layout')
|
||||
|
||||
# create the file structure
|
||||
pcgts = self.pcgts if self.pcgts else create_page_xml(self.image_filename, self.height_org, self.width_org)
|
||||
page = pcgts.get_Page()
|
||||
page.set_Border(BorderType(Coords=CoordsType(points=self.calculate_page_coords(cont_page))))
|
||||
|
||||
counter = EynollahIdCounter()
|
||||
_counter_marginals = EynollahIdCounter(region_idx=len(order_of_texts))
|
||||
id_of_marginalia = [_counter_marginals.next_region_id for _ in found_polygons_marginals]
|
||||
xml_reading_order(page, order_of_texts, id_of_marginalia)
|
||||
|
||||
for mm in range(len(found_polygons_text_region)):
|
||||
textregion = TextRegionType(id=counter.next_region_id, type_='paragraph',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(found_polygons_text_region[mm], page_coord), conf=conf_contours_textregion[mm]))
|
||||
page.add_TextRegion(textregion)
|
||||
|
||||
if ocr_all_textlines:
|
||||
ocr_textlines = ocr_all_textlines[mm]
|
||||
else:
|
||||
ocr_textlines = None
|
||||
self.serialize_lines_in_region(textregion, all_found_textline_polygons, mm, page_coord, all_box_coord, slopes, counter, ocr_textlines)
|
||||
|
||||
self.logger.debug('len(found_polygons_text_region_h) %s', len(found_polygons_text_region_h))
|
||||
for mm in range(len(found_polygons_text_region_h)):
|
||||
textregion = TextRegionType(id=counter.next_region_id, type_='header',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(found_polygons_text_region_h[mm], page_coord)))
|
||||
page.add_TextRegion(textregion)
|
||||
|
||||
if ocr_all_textlines:
|
||||
ocr_textlines = ocr_all_textlines[mm]
|
||||
else:
|
||||
ocr_textlines = None
|
||||
self.serialize_lines_in_region(textregion, all_found_textline_polygons_h, mm, page_coord, all_box_coord_h, slopes_h, counter, ocr_textlines)
|
||||
|
||||
for mm in range(len(found_polygons_marginals)):
|
||||
marginal = TextRegionType(id=counter.next_region_id, type_='marginalia',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(found_polygons_marginals[mm], page_coord)))
|
||||
page.add_TextRegion(marginal)
|
||||
self.serialize_lines_in_marginal(marginal, all_found_textline_polygons_marginals, mm, page_coord, all_box_coord_marginals, slopes_marginals, counter)
|
||||
|
||||
for mm in range(len(found_polygons_drop_capitals)):
|
||||
dropcapital = TextRegionType(id=counter.next_region_id, type_='drop-capital',
|
||||
Coords=CoordsType(points=self.calculate_polygon_coords(found_polygons_drop_capitals[mm], page_coord)))
|
||||
page.add_TextRegion(dropcapital)
|
||||
###all_box_coord_drop = None
|
||||
###slopes_drop = None
|
||||
###self.serialize_lines_in_dropcapital(dropcapital, [found_polygons_drop_capitals[mm]], mm, page_coord, all_box_coord_drop, slopes_drop, counter, ocr_all_textlines_textregion=None)
|
||||
|
||||
for mm in range(len(found_polygons_text_region_img)):
|
||||
page.add_ImageRegion(ImageRegionType(id=counter.next_region_id, Coords=CoordsType(points=self.calculate_polygon_coords(found_polygons_text_region_img[mm], page_coord))))
|
||||
|
||||
for mm in range(len(polygons_lines_to_be_written_in_xml)):
|
||||
page.add_SeparatorRegion(ImageRegionType(id=counter.next_region_id, Coords=CoordsType(points=self.calculate_polygon_coords(polygons_lines_to_be_written_in_xml[mm], [0 , 0, 0, 0]))))
|
||||
|
||||
for mm in range(len(found_polygons_tables)):
|
||||
page.add_TableRegion(TableRegionType(id=counter.next_region_id, Coords=CoordsType(points=self.calculate_polygon_coords(found_polygons_tables[mm], page_coord))))
|
||||
|
||||
return pcgts
|
||||
|
||||
def calculate_polygon_coords(self, contour, page_coord):
|
||||
self.logger.debug('enter calculate_polygon_coords')
|
||||
coords = ''
|
||||
for point in contour:
|
||||
if len(point) != 2:
|
||||
point = point[0]
|
||||
point_x = point[0]
|
||||
point_y = point[1]
|
||||
if not skip_layout_reading_order:
|
||||
point_x += page_coord[2]
|
||||
point_y += page_coord[0]
|
||||
point_x = int(point_x / self.scale_x)
|
||||
point_y = int(point_y / self.scale_y)
|
||||
coords += str(point_x) + ',' + str(point_y) + ' '
|
||||
for value_bbox in contour:
|
||||
if len(value_bbox) == 2:
|
||||
coords += str(int((value_bbox[0] + page_coord[2]) / self.scale_x))
|
||||
coords += ','
|
||||
coords += str(int((value_bbox[1] + page_coord[0]) / self.scale_y))
|
||||
else:
|
||||
coords += str(int((value_bbox[0][0] + page_coord[2]) / self.scale_x))
|
||||
coords += ','
|
||||
coords += str(int((value_bbox[0][1] + page_coord[0]) / self.scale_y))
|
||||
coords=coords + ' '
|
||||
return coords[:-1]
|
||||
|
||||
|
|
|
|||
|
|
@ -1,47 +0,0 @@
|
|||
from typing import List
|
||||
import pytest
|
||||
import logging
|
||||
|
||||
from click.testing import CliRunner, Result
|
||||
from eynollah.cli import main as eynollah_cli
|
||||
|
||||
|
||||
@pytest.fixture
|
||||
def run_eynollah_ok_and_check_logs(
|
||||
pytestconfig,
|
||||
caplog,
|
||||
model_dir,
|
||||
eynollah_subcommands,
|
||||
eynollah_log_filter,
|
||||
):
|
||||
"""
|
||||
Generates a Click Runner for `cli`, injects model_path and logging level
|
||||
to `args`, runs the command and checks whether the logs generated contain
|
||||
every fragment in `expected_logs`
|
||||
"""
|
||||
|
||||
def _run_click_ok_logs(
|
||||
subcommand: 'str',
|
||||
args: List[str],
|
||||
expected_logs: List[str],
|
||||
) -> Result:
|
||||
assert subcommand in eynollah_subcommands, f'subcommand {subcommand} must be one of {eynollah_subcommands}'
|
||||
args = [
|
||||
'-m', model_dir,
|
||||
subcommand,
|
||||
*args
|
||||
]
|
||||
if pytestconfig.getoption('verbose') > 0:
|
||||
args = ['-l', 'DEBUG'] + args
|
||||
caplog.set_level(logging.INFO)
|
||||
runner = CliRunner()
|
||||
with caplog.filtering(eynollah_log_filter):
|
||||
result = runner.invoke(eynollah_cli, args, catch_exceptions=False)
|
||||
assert result.exit_code == 0, result.stdout
|
||||
if expected_logs:
|
||||
logmsgs = [logrec.message for logrec in caplog.records]
|
||||
assert any(logmsg.startswith(needle) for needle in expected_logs for logmsg in logmsgs), f'{expected_logs} not in {logmsgs}'
|
||||
return result
|
||||
|
||||
return _run_click_ok_logs
|
||||
|
||||
|
|
@ -1,53 +0,0 @@
|
|||
import pytest
|
||||
from PIL import Image
|
||||
|
||||
@pytest.mark.parametrize(
|
||||
"options",
|
||||
[
|
||||
[], # defaults
|
||||
["--no-patches"],
|
||||
], ids=str)
|
||||
def test_run_eynollah_binarization_filename(
|
||||
tmp_path,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
resources_dir,
|
||||
options,
|
||||
):
|
||||
infile = resources_dir / '2files/kant_aufklaerung_1784_0020.tif'
|
||||
outfile = tmp_path / 'kant_aufklaerung_1784_0020.png'
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'binarization',
|
||||
[
|
||||
'-i', str(infile),
|
||||
'-o', str(outfile),
|
||||
] + options,
|
||||
[
|
||||
'Predicting'
|
||||
]
|
||||
)
|
||||
assert outfile.exists()
|
||||
with Image.open(infile) as original_img:
|
||||
original_size = original_img.size
|
||||
with Image.open(outfile) as binarized_img:
|
||||
binarized_size = binarized_img.size
|
||||
assert original_size == binarized_size
|
||||
|
||||
def test_run_eynollah_binarization_directory(
|
||||
tmp_path,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
resources_dir,
|
||||
image_resources,
|
||||
):
|
||||
outdir = tmp_path
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'binarization',
|
||||
[
|
||||
'-di', str(resources_dir / '2files'),
|
||||
'-o', str(outdir),
|
||||
],
|
||||
[
|
||||
f'Predicting {image_resources[0].name}',
|
||||
f'Predicting {image_resources[1].name}',
|
||||
]
|
||||
)
|
||||
assert len(list(outdir.iterdir())) == 2
|
||||
|
|
@ -1,52 +0,0 @@
|
|||
import pytest
|
||||
from PIL import Image
|
||||
|
||||
@pytest.mark.parametrize(
|
||||
"options",
|
||||
[
|
||||
[], # defaults
|
||||
["-sos"],
|
||||
], ids=str)
|
||||
def test_run_eynollah_enhancement_filename(
|
||||
tmp_path,
|
||||
resources_dir,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
options,
|
||||
):
|
||||
infile = resources_dir / '2files/kant_aufklaerung_1784_0020.tif'
|
||||
outfile = tmp_path / 'kant_aufklaerung_1784_0020.png'
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'enhancement',
|
||||
[
|
||||
'-i', str(infile),
|
||||
'-o', str(outfile.parent),
|
||||
] + options,
|
||||
[
|
||||
'Image was enhanced',
|
||||
]
|
||||
)
|
||||
with Image.open(infile) as original_img:
|
||||
original_size = original_img.size
|
||||
with Image.open(outfile) as enhanced_img:
|
||||
enhanced_size = enhanced_img.size
|
||||
assert (original_size == enhanced_size) == ("-sos" in options)
|
||||
|
||||
def test_run_eynollah_enhancement_directory(
|
||||
tmp_path,
|
||||
resources_dir,
|
||||
image_resources,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
):
|
||||
outdir = tmp_path
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'enhancement',
|
||||
[
|
||||
'-di', str(resources_dir/ '2files'),
|
||||
'-o', str(outdir),
|
||||
],
|
||||
[
|
||||
f'Image {image_resources[0]} was enhanced',
|
||||
f'Image {image_resources[1]} was enhanced',
|
||||
]
|
||||
)
|
||||
assert len(list(outdir.iterdir())) == 2
|
||||
|
|
@ -1,119 +0,0 @@
|
|||
import pytest
|
||||
from ocrd_modelfactory import page_from_file
|
||||
from ocrd_models.constants import NAMESPACES as NS
|
||||
|
||||
@pytest.mark.parametrize(
|
||||
"options",
|
||||
[
|
||||
[], # defaults
|
||||
#["--allow_scaling", "--curved-line"],
|
||||
["--allow_scaling", "--curved-line", "--full-layout"],
|
||||
["--allow_scaling", "--curved-line", "--full-layout", "--reading_order_machine_based"],
|
||||
# -ep ...
|
||||
# -eoi ...
|
||||
# --skip_layout_and_reading_order
|
||||
], ids=str)
|
||||
def test_run_eynollah_layout_filename(
|
||||
tmp_path,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
resources_dir,
|
||||
options,
|
||||
):
|
||||
infile = resources_dir / '2files/kant_aufklaerung_1784_0020.tif'
|
||||
outfile = tmp_path / 'kant_aufklaerung_1784_0020.xml'
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'layout',
|
||||
[
|
||||
'-i', str(infile),
|
||||
'-o', str(outfile.parent),
|
||||
] + options,
|
||||
[
|
||||
str(infile)
|
||||
]
|
||||
)
|
||||
assert outfile.exists()
|
||||
tree = page_from_file(str(outfile)).etree
|
||||
regions = tree.xpath("//page:TextRegion", namespaces=NS)
|
||||
assert len(regions) >= 2, "result is inaccurate"
|
||||
regions = tree.xpath("//page:SeparatorRegion", namespaces=NS)
|
||||
assert len(regions) >= 2, "result is inaccurate"
|
||||
lines = tree.xpath("//page:TextLine", namespaces=NS)
|
||||
assert len(lines) == 31, "result is inaccurate" # 29 paragraph lines, 1 page and 1 catch-word line
|
||||
|
||||
@pytest.mark.parametrize(
|
||||
"options",
|
||||
[
|
||||
["--tables"],
|
||||
["--tables", "--full-layout"],
|
||||
], ids=str)
|
||||
def test_run_eynollah_layout_filename2(
|
||||
tmp_path,
|
||||
resources_dir,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
options,
|
||||
):
|
||||
infile = resources_dir / '2files/euler_rechenkunst01_1738_0025.tif'
|
||||
outfile = tmp_path / 'euler_rechenkunst01_1738_0025.xml'
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'layout',
|
||||
[
|
||||
'-i', str(infile),
|
||||
'-o', str(outfile.parent),
|
||||
] + options,
|
||||
[
|
||||
str(infile)
|
||||
]
|
||||
)
|
||||
assert outfile.exists()
|
||||
tree = page_from_file(str(outfile)).etree
|
||||
regions = tree.xpath("//page:TextRegion", namespaces=NS)
|
||||
assert len(regions) >= 2, "result is inaccurate"
|
||||
regions = tree.xpath("//page:TableRegion", namespaces=NS)
|
||||
# model/decoding is not very precise, so (depending on mode) we can get fractures/splits/FP
|
||||
assert len(regions) >= 1, "result is inaccurate"
|
||||
regions = tree.xpath("//page:SeparatorRegion", namespaces=NS)
|
||||
assert len(regions) >= 2, "result is inaccurate"
|
||||
lines = tree.xpath("//page:TextLine", namespaces=NS)
|
||||
assert len(lines) >= 2, "result is inaccurate" # mostly table (if detected correctly), but 1 page and 1 catch-word line
|
||||
|
||||
def test_run_eynollah_layout_directory(
|
||||
tmp_path,
|
||||
resources_dir,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
):
|
||||
outdir = tmp_path
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'layout',
|
||||
[
|
||||
'-di', str(resources_dir / '2files'),
|
||||
'-o', str(outdir),
|
||||
],
|
||||
[
|
||||
'Job done in',
|
||||
'All jobs done in',
|
||||
]
|
||||
)
|
||||
assert len(list(outdir.iterdir())) == 2
|
||||
|
||||
# def test_run_eynollah_layout_marginalia(
|
||||
# tmp_path,
|
||||
# resources_dir,
|
||||
# run_eynollah_ok_and_check_logs,
|
||||
# ):
|
||||
# outdir = tmp_path
|
||||
# outfile = outdir / 'estor_rechtsgelehrsamkeit02_1758_0880_800px.xml'
|
||||
# run_eynollah_ok_and_check_logs(
|
||||
# 'layout',
|
||||
# [
|
||||
# '-i', str(resources_dir / 'estor_rechtsgelehrsamkeit02_1758_0880_800px.jpg'),
|
||||
# '-o', str(outdir),
|
||||
# ],
|
||||
# [
|
||||
# 'Job done in',
|
||||
# 'All jobs done in',
|
||||
# ]
|
||||
# )
|
||||
# assert outfile.exists()
|
||||
# tree = page_from_file(str(outfile)).etree
|
||||
# regions = tree.xpath('//page:TextRegion[type="marginalia"]', namespaces=NS)
|
||||
# assert len(regions) == 5, "expected 5 marginalia regions"
|
||||
|
|
@ -1,47 +0,0 @@
|
|||
from ocrd_modelfactory import page_from_file
|
||||
from ocrd_models.constants import NAMESPACES as NS
|
||||
|
||||
def test_run_eynollah_mbreorder_filename(
|
||||
tmp_path,
|
||||
resources_dir,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
):
|
||||
infile = resources_dir / '2files/kant_aufklaerung_1784_0020.xml'
|
||||
outfile = tmp_path /'kant_aufklaerung_1784_0020.xml'
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'machine-based-reading-order',
|
||||
[
|
||||
'-i', str(infile),
|
||||
'-o', str(outfile.parent),
|
||||
],
|
||||
[
|
||||
# FIXME: mbreorder has no logging!
|
||||
]
|
||||
)
|
||||
assert outfile.exists()
|
||||
#in_tree = page_from_file(str(infile)).etree
|
||||
#in_order = in_tree.xpath("//page:OrderedGroup//@regionRef", namespaces=NS)
|
||||
out_tree = page_from_file(str(outfile)).etree
|
||||
out_order = out_tree.xpath("//page:OrderedGroup//@regionRef", namespaces=NS)
|
||||
#assert len(out_order) >= 2, "result is inaccurate"
|
||||
#assert in_order != out_order
|
||||
assert out_order == ['r_1_1', 'r_2_1', 'r_2_2', 'r_2_3']
|
||||
|
||||
def test_run_eynollah_mbreorder_directory(
|
||||
tmp_path,
|
||||
resources_dir,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
):
|
||||
outdir = tmp_path
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'machine-based-reading-order',
|
||||
[
|
||||
'-di', str(resources_dir / '2files'),
|
||||
'-o', str(outdir),
|
||||
],
|
||||
[
|
||||
# FIXME: mbreorder has no logging!
|
||||
]
|
||||
)
|
||||
assert len(list(outdir.iterdir())) == 2
|
||||
|
||||
|
|
@ -1,64 +0,0 @@
|
|||
import pytest
|
||||
from ocrd_modelfactory import page_from_file
|
||||
from ocrd_models.constants import NAMESPACES as NS
|
||||
|
||||
@pytest.mark.parametrize(
|
||||
"options",
|
||||
[
|
||||
["-trocr"],
|
||||
[], # defaults
|
||||
["-doit", #str(outrenderfile.parent)],
|
||||
],
|
||||
], ids=str)
|
||||
def test_run_eynollah_ocr_filename(
|
||||
tmp_path,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
resources_dir,
|
||||
options,
|
||||
):
|
||||
infile = resources_dir / '2files/kant_aufklaerung_1784_0020.tif'
|
||||
outfile = tmp_path / 'kant_aufklaerung_1784_0020.xml'
|
||||
outrenderfile = tmp_path / 'render' / 'kant_aufklaerung_1784_0020.png'
|
||||
outrenderfile.parent.mkdir()
|
||||
if "-doit" in options:
|
||||
options.insert(options.index("-doit") + 1, str(outrenderfile.parent))
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'ocr',
|
||||
[
|
||||
'-i', str(infile),
|
||||
'-dx', str(infile.parent),
|
||||
'-o', str(outfile.parent),
|
||||
] + options,
|
||||
[
|
||||
# FIXME: ocr has no logging!
|
||||
]
|
||||
)
|
||||
assert outfile.exists()
|
||||
if "-doit" in options:
|
||||
assert outrenderfile.exists()
|
||||
#in_tree = page_from_file(str(infile)).etree
|
||||
#in_order = in_tree.xpath("//page:OrderedGroup//@regionRef", namespaces=NS)
|
||||
out_tree = page_from_file(str(outfile)).etree
|
||||
out_texts = out_tree.xpath("//page:TextLine/page:TextEquiv[last()]/page:Unicode/text()", namespaces=NS)
|
||||
assert len(out_texts) >= 2, ("result is inaccurate", out_texts)
|
||||
assert sum(map(len, out_texts)) > 100, ("result is inaccurate", out_texts)
|
||||
|
||||
def test_run_eynollah_ocr_directory(
|
||||
tmp_path,
|
||||
run_eynollah_ok_and_check_logs,
|
||||
resources_dir,
|
||||
):
|
||||
outdir = tmp_path
|
||||
run_eynollah_ok_and_check_logs(
|
||||
'ocr',
|
||||
[
|
||||
'-di', str(resources_dir / '2files'),
|
||||
'-dx', str(resources_dir / '2files'),
|
||||
'-o', str(outdir),
|
||||
],
|
||||
[
|
||||
# FIXME: ocr has no logging!
|
||||
]
|
||||
)
|
||||
assert len(list(outdir.iterdir())) == 2
|
||||
|
||||
|
|
@ -1,37 +0,0 @@
|
|||
from glob import glob
|
||||
import os
|
||||
import pytest
|
||||
from pathlib import Path
|
||||
|
||||
|
||||
@pytest.fixture()
|
||||
def tests_dir():
|
||||
return Path(__file__).parent.resolve()
|
||||
|
||||
@pytest.fixture()
|
||||
def model_dir(tests_dir):
|
||||
return os.environ.get('EYNOLLAH_MODELS_DIR', str(tests_dir.joinpath('..').resolve()))
|
||||
|
||||
@pytest.fixture()
|
||||
def resources_dir(tests_dir):
|
||||
return tests_dir / 'resources'
|
||||
|
||||
@pytest.fixture()
|
||||
def image_resources(resources_dir):
|
||||
return [Path(x) for x in glob(str(resources_dir / '2files/*.tif'))]
|
||||
|
||||
@pytest.fixture()
|
||||
def eynollah_log_filter():
|
||||
return lambda logrec: logrec.name.startswith('eynollah')
|
||||
|
||||
@pytest.fixture
|
||||
def eynollah_subcommands():
|
||||
return [
|
||||
'binarization',
|
||||
'layout',
|
||||
'ocr',
|
||||
'enhancement',
|
||||
'machine-based-reading-order',
|
||||
'models',
|
||||
]
|
||||
|
||||
File diff suppressed because it is too large
Load diff
File diff suppressed because it is too large
Load diff
Binary file not shown.
|
Before Width: | Height: | Size: 221 KiB |
|
|
@ -1,235 +0,0 @@
|
|||
<?xml version="1.0" encoding="UTF-8"?>
|
||||
<pc:PcGts xmlns:pc="http://schema.primaresearch.org/PAGE/gts/pagecontent/2019-07-15" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://schema.primaresearch.org/PAGE/gts/pagecontent/2019-07-15 http://schema.primaresearch.org/PAGE/gts/pagecontent/2019-07-15/pagecontent.xsd">
|
||||
<pc:Metadata>
|
||||
<pc:Creator>SBB_QURATOR</pc:Creator>
|
||||
<pc:Created>2025-10-30T16:38:21.180191</pc:Created>
|
||||
<pc:LastChange>2025-10-30T16:38:21.180191</pc:LastChange>
|
||||
</pc:Metadata>
|
||||
<pc:Page imageFilename="/home/vahid/Downloads/estor_rechtsgelehrsamkeit02_1758_0880_800px.jpg" imageWidth="800" imageHeight="1376" readingDirection="left-to-right" textLineOrder="top-to-bottom">
|
||||
<pc:Border>
|
||||
<pc:Coords points="780,40 780,42 780,43 736,43 736,45 735,46 675,46 675,48 674,49 627,49 627,52 626,52 570,52 570,55 569,56 530,56 530,57 529,58 472,58 472,60 471,61 296,61 296,60 296,58 66,58 65,57 65,56 50,56 50,57 49,58 47,58 47,60 46,61 44,61 44,64 46,64 47,64 47,79 48,79 48,80 48,162 48,163 47,163 47,340 48,340 48,341 48,364 48,365 47,365 47,444 48,444 48,445 48,1052 49,1052 50,1053 50,1182 51,1182 52,1183 52,1332 74,1332 74,1330 75,1329 76,1329 76,1330 76,1332 78,1332 78,1330 79,1329 80,1329 80,1330 80,1332 87,1332 87,1330 88,1329 272,1329 272,1327 273,1326 380,1326 380,1324 380,1323 401,1323 401,1320 402,1320 423,1320 423,1318 424,1317 444,1317 444,1315 444,1314 458,1314 458,1312 459,1311 472,1311 472,1308 473,1308 512,1308 512,1305 512,1304 569,1304 569,1302 570,1301 601,1301 601,1300 602,1299 624,1299 624,1296 625,1296 644,1296 644,1293 644,1292 708,1292 708,1290 709,1289 790,1289 790,1287 791,1286 792,1286 792,1272 793,1271 794,1271 794,1152 793,1152 792,1151 792,1128 793,1127 794,1127 794,1108 793,1108 792,1108 792,1103 793,1102 794,1102 794,1075 793,1075 792,1074 792,829 793,828 794,828 794,820 793,820 792,820 792,789 791,789 790,788 790,786 791,785 792,785 792,784 791,784 790,783 790,697 791,696 792,696 792,685 791,685 790,684 790,632 789,632 788,632 788,525 788,525 787,524 787,212 786,212 785,211 785,43 784,43 783,42 783,40"/>
|
||||
</pc:Border>
|
||||
<pc:ReadingOrder>
|
||||
<pc:OrderedGroup id="ro357564684568544579089">
|
||||
<pc:RegionRefIndexed index="0" regionRef="region_0013"/>
|
||||
<pc:RegionRefIndexed index="1" regionRef="region_0014"/>
|
||||
<pc:RegionRefIndexed index="2" regionRef="region_0015"/>
|
||||
<pc:RegionRefIndexed index="3" regionRef="region_0016"/>
|
||||
<pc:RegionRefIndexed index="4" regionRef="region_0017"/>
|
||||
<pc:RegionRefIndexed index="5" regionRef="region_0012"/>
|
||||
<pc:RegionRefIndexed index="6" regionRef="region_0010"/>
|
||||
<pc:RegionRefIndexed index="7" regionRef="region_0006"/>
|
||||
<pc:RegionRefIndexed index="8" regionRef="region_0011"/>
|
||||
<pc:RegionRefIndexed index="9" regionRef="region_0003"/>
|
||||
<pc:RegionRefIndexed index="10" regionRef="region_0009"/>
|
||||
<pc:RegionRefIndexed index="11" regionRef="region_0005"/>
|
||||
<pc:RegionRefIndexed index="12" regionRef="region_0007"/>
|
||||
<pc:RegionRefIndexed index="13" regionRef="region_0004"/>
|
||||
<pc:RegionRefIndexed index="14" regionRef="region_0008"/>
|
||||
<pc:RegionRefIndexed index="15" regionRef="region_0002"/>
|
||||
<pc:RegionRefIndexed index="16" regionRef="region_0001"/>
|
||||
</pc:OrderedGroup>
|
||||
</pc:ReadingOrder>
|
||||
<pc:TextRegion id="region_0001" orientation="1.241379310344827" type="paragraph">
|
||||
<pc:Coords points="743,1172 740,1172 740,1172 716,1170 714,1172 709,1172 707,1172 702,1172 697,1177 697,1175 692,1175 690,1175 671,1175 668,1180 668,1177 668,1177 664,1180 664,1180 664,1201 666,1204 664,1206 668,1208 666,1208 666,1206 668,1211 668,1211 685,1213 690,1208 690,1211 690,1211 692,1208 697,1211 700,1208 704,1208 709,1208 714,1208 719,1204 719,1206 731,1206 733,1204 733,1204 748,1206 750,1204 750,1204 748,1204 752,1206 755,1201 755,1201 752,1204 757,1201 755,1201 760,1194 760,1196 757,1196 760,1175 757,1172 757,1172 755,1170 755,1170 757,1172" conf="0.21158782745235"/>
|
||||
<pc:TextLine id="region_0001_line_0001">
|
||||
<pc:Coords points="751,1167 747,1169 745,1168 741,1170 744,1168 740,1171 736,1169 732,1172 735,1170 731,1172 731,1171 727,1173 728,1172 724,1174 723,1172 719,1175 719,1173 715,1176 716,1174 712,1176 712,1175 708,1177 688,1176 684,1178 675,1176 671,1179 672,1177 668,1180 671,1178 667,1180 665,1185 666,1183 664,1194 667,1198 665,1204 668,1208 666,1207 676,1212 680,1209 681,1211 685,1208 684,1210 688,1207 688,1208 692,1206 711,1208 715,1205 714,1207 718,1204 718,1206 722,1204 720,1205 718,1204 730,1206 734,1204 732,1205 736,1203 734,1204 738,1202 738,1204 742,1201 740,1203 744,1200 741,1202 746,1199 746,1200 750,1198 748,1200 752,1196 752,1198 757,1194 759,1190 758,1192 760,1188 759,1191 760,1183 758,1179 760,1174 754,1169 756,1170 758,1172 756,1168 751,1167"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0002" orientation="1.241379310344827" type="paragraph">
|
||||
<pc:Coords points="724,1086 719,1084 700,1084 697,1086 690,1084 685,1088 685,1088 680,1086 678,1088 671,1088 666,1088 652,1088 649,1093 649,1091 642,1091 640,1091 628,1091 623,1096 623,1093 623,1093 623,1093 623,1100 625,1100 625,1100 611,1093 606,1096 606,1096 592,1093 587,1096 587,1096 589,1096 568,1093 563,1096 560,1096 556,1100 556,1100 556,1100 558,1098 558,1100 558,1100 558,1100 558,1100 551,1096 546,1096 534,1096 529,1098 520,1096 515,1100 515,1100 498,1098 496,1100 500,1100 469,1098 467,1100 342,1100 340,1100 330,1100 330,1100 328,1103 328,1103 330,1103 330,1103 323,1100 323,1100 323,1100 320,1105 323,1105 323,1105 308,1100 308,1100 294,1100 289,1100 289,1100 287,1129 287,1129 287,1129 284,1132 284,1132 284,1129 287,1129 287,1132 263,1129 258,1132 258,1132 258,1129 260,1129 260,1132 253,1129 251,1132 251,1132 251,1129 248,1132 246,1158 248,1160 248,1160 248,1163 248,1163 282,1165 287,1160 287,1163 287,1163 284,1163 284,1163 289,1165 292,1163 292,1165 289,1165 342,1165 344,1160 344,1163 352,1163 352,1163 349,1160 356,1165 356,1165 378,1165 380,1160 380,1163 404,1163 407,1160 436,1163 433,1163 448,1163 450,1160 457,1163 462,1160 464,1160 469,1160 467,1160 469,1156 469,1158 472,1134 472,1136 469,1136 472,1136 474,1132 474,1134 479,1134 484,1132 481,1134 484,1132 498,1132 500,1129 500,1129 508,1132 508,1132 508,1132 510,1127 508,1127 508,1127 515,1132 520,1129 541,1129 546,1129 575,1129 577,1124 577,1127 611,1127 613,1124 623,1127 625,1124 637,1124 642,1124 652,1124 654,1120 654,1122 656,1122 659,1120 659,1120 664,1122 666,1120 680,1120 685,1117 685,1117 685,1120 683,1120 683,1117 707,1120 712,1117 712,1117 721,1120 719,1120 719,1117 728,1120 733,1117 733,1117 731,1120 733,1117 736,1117 738,1117 738,1117 740,1112 736,1110 736,1110 738,1112 738,1112 738,1112 736,1110 736,1110 736,1110 736,1110 736,1110 733,1110 733,1110 745,1115 750,1112 752,1115 757,1112 757,1108 757,1110 760,1105 760,1105 757,1108 760,1098 760,1100 762,1096 760,1093 760,1096 762,1088 760,1084 760,1086" conf="0.285657570399826"/>
|
||||
<pc:TextLine id="region_0002_line_0001">
|
||||
<pc:Coords points="741,1081 737,1084 735,1082 731,1084 719,1083 715,1085 712,1084 714,1084 708,1083 711,1084 702,1082 697,1085 696,1089 696,1087 696,1085 692,1088 688,1086 684,1088 687,1087 682,1092 683,1090 684,1088 680,1091 680,1089 676,1092 667,1090 663,1092 644,1091 640,1093 639,1092 634,1095 626,1093 626,1093 626,1093 624,1098 624,1098 624,1098 601,1093 597,1096 603,1095 579,1093 581,1094 573,1092 569,1095 571,1093 567,1096 566,1094 562,1096 560,1095 555,1099 556,1097 559,1098 555,1096 551,1099 506,1097 502,1100 499,1098 495,1100 464,1099 466,1100 380,1098 375,1101 375,1101 374,1103 376,1101 379,1102 379,1102 360,1100 356,1102 348,1100 344,1103 342,1101 344,1102 346,1104 340,1100 343,1100 330,1099 330,1099 326,1102 328,1100 330,1101 330,1101 323,1099 323,1099 323,1099 320,1103 320,1103 320,1103 308,1099 310,1100 293,1098 289,1100 292,1099 288,1101 286,1108 287,1105 285,1117 288,1121 286,1122 288,1126 287,1126 289,1130 288,1128 293,1132 291,1132 299,1133 296,1132 311,1134 315,1132 318,1133 322,1131 326,1132 330,1130 341,1132 346,1126 345,1128 374,1130 372,1129 379,1128 373,1129 378,1131 376,1131 375,1129 391,1134 395,1132 395,1133 399,1131 396,1132 401,1126 401,1128 428,1129 432,1127 507,1128 511,1126 509,1128 507,1127 512,1128 516,1126 557,1128 561,1125 572,1127 576,1124 591,1126 595,1124 612,1125 616,1123 624,1124 628,1122 626,1124 630,1121 628,1123 632,1120 645,1122 649,1120 657,1121 661,1119 665,1120 669,1118 675,1120 679,1117 682,1119 686,1116 694,1118 698,1116 720,1117 724,1115 727,1116 731,1114 729,1116 734,1110 733,1112 735,1108 735,1109 732,1111 736,1108 736,1110 740,1108 739,1109 743,1107 741,1108 745,1106 744,1108 748,1105 746,1107 750,1104 748,1106 752,1104 749,1105 747,1104 751,1106 756,1102 758,1096 757,1099 759,1093 758,1096 760,1087 757,1083 741,1081"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0002_line_0002">
|
||||
<pc:Coords points="412,1126 408,1128 400,1127 395,1130 397,1128 393,1131 394,1129 390,1132 387,1130 383,1132 376,1131 379,1132 373,1130 376,1131 360,1129 355,1132 356,1131 352,1133 353,1132 356,1132 350,1131 352,1132 348,1130 348,1129 339,1128 335,1130 327,1128 323,1131 320,1129 316,1132 308,1130 304,1132 296,1131 298,1132 290,1130 292,1131 292,1131 292,1131 292,1131 288,1134 288,1134 288,1134 291,1133 284,1128 286,1128 262,1127 262,1127 258,1130 258,1128 260,1129 256,1128 252,1130 253,1128 249,1131 252,1129 248,1132 250,1130 245,1158 248,1162 246,1160 248,1164 247,1162 283,1167 283,1167 287,1163 286,1164 284,1164 288,1165 292,1163 312,1164 316,1162 328,1164 325,1163 335,1164 332,1164 336,1165 340,1163 338,1164 342,1162 341,1164 345,1161 345,1163 349,1160 351,1156 350,1159 348,1160 352,1158 358,1160 362,1157 405,1159 403,1158 409,1160 407,1159 411,1160 408,1160 414,1161 412,1160 421,1162 425,1160 435,1161 432,1160 444,1162 449,1159 451,1155 452,1156 459,1157 463,1155 463,1156 467,1154 464,1156 469,1148 467,1144 461,1142 462,1141 464,1138 460,1133 431,1132 433,1132 428,1131 430,1132 423,1130 425,1131 421,1129 424,1130 425,1132 423,1128 419,1127 421,1128 412,1126"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0003" orientation="1.241379310344827" type="paragraph">
|
||||
<pc:Coords points="738,520 731,520 728,520 726,520 726,520 721,522 714,520 712,524 712,522 700,522 695,527 695,524 635,524 635,524 637,524 630,522 628,527 628,524 620,524 616,524 606,524 601,529 601,527 572,527 570,527 568,527 568,527 568,527 565,529 565,529 565,529 563,527 563,527 553,527 551,527 522,527 517,529 390,527 388,532 388,532 373,529 371,532 361,532 364,532 349,529 347,532 335,532 332,532 335,532 335,532 313,532 313,532 292,529 289,532 287,536 289,539 289,551 289,548 287,553 289,558 289,558 289,556 287,558 287,558 287,560 287,560 287,558 284,560 282,560 280,560 272,560 268,565 268,563 263,563 260,565 260,565 258,563 260,563 253,563 251,565 251,563 248,568 248,568 248,577 248,575 246,599 246,596 246,618 248,620 253,620 253,620 349,623 354,620 354,620 354,620 359,623 361,620 385,620 388,620 397,620 400,616 400,618 400,618 402,616 402,618 404,594 404,596 404,592 407,594 404,594 409,592 409,594 407,594 414,594 419,592 428,594 431,592 433,592 436,589 436,589 520,592 522,589 529,589 534,589 551,589 548,589 556,589 558,589 558,589 558,589 611,589 608,589 623,592 625,589 659,589 656,589 695,592 697,589 697,589 700,589 700,589 702,587 709,589 712,584 714,584 712,587 714,584 714,584 719,584 719,584 721,580 721,582 726,582 724,582 724,582 726,584 726,584 728,584 733,584 738,584 740,580 740,582 750,582 752,582 755,582 757,577 757,580 757,577 757,577 757,580 760,575 760,575 762,570 760,568 760,539 760,541 762,527 760,524 760,524 757,522 757,522 760,524 752,520 755,520 748,520 750,520 738,520 740,520" conf="0.410852900020834"/>
|
||||
<pc:TextLine id="region_0003_line_0001">
|
||||
<pc:Coords points="735,516 731,519 732,517 728,523 728,520 727,524 728,522 728,520 724,523 720,521 716,524 716,522 712,524 675,523 671,525 640,524 643,524 637,523 640,524 634,522 636,523 638,525 631,520 626,526 627,524 624,522 620,524 617,523 613,525 613,524 609,526 604,524 600,527 598,525 600,526 587,524 583,527 567,525 567,525 567,525 564,528 567,529 567,529 553,525 549,528 544,526 540,528 543,527 539,529 540,528 536,530 533,528 529,531 513,529 509,532 502,530 504,531 500,529 501,529 448,528 451,528 440,527 442,528 428,526 424,528 404,527 400,529 395,528 391,530 376,528 372,531 370,529 366,532 332,530 335,531 312,529 315,530 308,531 297,529 293,532 296,530 292,532 290,536 291,534 289,546 292,550 290,552 292,556 291,556 293,560 318,562 322,560 326,561 330,559 359,560 363,558 365,560 363,559 372,560 372,560 370,559 374,560 372,560 370,557 373,562 388,564 392,561 410,563 414,560 416,556 416,558 420,560 424,557 464,559 468,556 516,558 514,557 521,559 520,559 526,560 530,558 528,560 533,556 531,558 535,556 533,557 537,555 548,556 552,554 591,556 588,555 592,556 591,556 616,558 614,557 619,559 623,556 623,558 627,556 624,557 630,553 667,555 671,552 687,554 684,553 695,555 699,552 704,554 708,552 706,553 710,551 708,552 712,550 717,552 721,549 727,551 731,548 732,550 736,548 736,549 740,547 740,548 744,546 743,548 747,545 746,547 751,544 752,540 752,541 750,543 754,540 753,542 758,536 757,539 759,533 758,536 760,526 757,522 740,520 741,520 743,522 740,518 735,516"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0003_line_0002">
|
||||
<pc:Coords points="719,548 715,550 717,548 712,552 711,556 711,554 716,553 712,552 708,554 709,552 705,555 705,553 701,556 697,554 693,556 696,555 692,557 694,556 696,556 687,555 683,557 673,556 676,556 666,555 668,556 664,554 667,555 663,553 665,554 640,552 642,553 636,552 639,552 635,551 637,552 632,550 628,552 630,551 626,553 628,552 624,554 625,552 621,555 582,553 578,556 576,554 579,555 574,553 570,556 572,554 574,555 568,553 564,556 560,554 562,555 535,553 531,556 532,554 528,556 528,555 524,557 522,556 524,556 514,555 510,557 512,556 508,558 509,556 505,559 500,557 496,560 472,558 468,560 464,559 465,559 456,557 452,560 455,558 451,560 439,559 435,561 414,560 416,560 412,559 415,560 397,558 400,559 393,557 389,560 386,558 382,560 379,559 381,560 371,558 373,559 358,557 354,560 351,558 347,560 316,559 319,560 293,558 289,560 284,559 280,562 278,566 278,564 280,563 276,565 267,564 263,566 252,564 248,568 246,587 248,591 247,590 259,595 263,592 268,594 272,592 280,593 284,591 300,592 298,592 312,593 316,591 314,592 319,587 319,588 320,590 324,588 341,589 345,587 365,588 369,586 398,588 396,587 419,588 417,588 416,588 425,592 429,590 431,585 430,588 432,584 431,586 475,588 479,585 507,587 504,586 513,588 511,587 521,588 525,586 523,588 527,585 528,581 528,583 534,584 538,582 575,584 579,581 604,583 601,582 608,584 612,581 614,583 612,582 617,584 616,584 614,582 616,586 629,588 633,585 652,587 649,586 666,588 670,585 676,587 673,586 685,588 690,582 689,584 691,580 691,582 705,584 709,581 712,583 716,580 716,582 720,580 721,581 725,579 728,580 732,578 731,580 735,577 736,579 740,576 744,578 748,576 751,577 755,575 752,576 757,573 759,566 758,568 760,562 757,558 759,560 753,556 756,556 752,555 753,555 755,556 752,551 740,549 736,552 732,550 735,551 728,549 731,550 723,548 725,549 719,548"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0003_line_0003">
|
||||
<pc:Coords points="360,584 356,586 357,584 353,587 342,585 338,588 330,586 326,588 322,587 318,589 309,588 305,590 305,588 308,589 283,588 278,593 279,591 277,596 277,594 257,592 253,595 250,593 246,596 244,615 269,620 273,617 321,619 319,618 326,617 320,618 324,620 324,620 338,622 342,620 349,621 353,619 379,620 383,618 384,614 384,616 389,617 393,615 396,616 400,614 402,608 400,604 392,602 394,603 396,600 393,596 395,597 392,593 392,593 392,593 369,594 369,594 369,594 371,596 372,590 369,585 360,584"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0004" orientation="1.241379310344827" type="paragraph">
|
||||
<pc:Coords points="740,940 738,940 738,940 733,937 731,940 719,940 714,940 712,940 707,942 692,940 690,944 690,944 685,942 683,944 671,944 668,944 630,944 628,949 628,947 599,947 596,947 553,947 551,952 551,949 532,949 532,949 529,952 529,949 532,949 532,949 527,949 524,952 524,952 515,949 517,949 517,952 467,949 464,952 464,952 448,949 450,949 450,952 438,949 440,949 440,949 431,947 426,952 426,949 412,949 407,952 407,952 385,949 383,952 352,952 349,952 304,952 306,952 294,952 292,952 294,952 289,956 289,956 292,954 289,959 289,956 287,978 287,976 287,980 287,980 287,980 284,980 284,980 282,985 282,983 280,983 275,983 270,983 268,985 263,983 260,985 258,985 260,985 251,983 248,988 248,988 248,1009 248,1007 246,1036 248,1038 248,1038 248,1040 368,1043 373,1040 373,1040 371,1040 378,1043 380,1040 414,1040 412,1040 443,1043 445,1040 481,1040 486,1040 520,1040 522,1036 522,1038 568,1038 570,1036 570,1036 599,1038 601,1036 601,1036 604,1033 604,1033 606,1024 604,1021 604,1012 606,1012 606,1009 606,1009 606,1012 608,1009 623,1009 620,1009 628,1012 630,1009 642,1009 644,1009 659,1009 664,1007 661,1009 664,1004 664,1007 671,1007 676,1004 695,1007 697,1004 697,1004 695,1004 697,1007 702,1004 700,1004 702,1004 709,1004 712,1000 712,1002 714,1002 719,1002 719,1002 721,1000 721,1002 726,1000 726,1000 728,997 728,997 728,1000 731,997 733,997 738,997 740,997 745,995 752,997 757,992 757,995 755,995 757,990 757,992 760,988 760,990 757,990 760,942 757,940 757,940 760,940" conf="0.530462165249498"/>
|
||||
<pc:TextLine id="region_0004_line_0001">
|
||||
<pc:Coords points="748,935 744,937 743,936 739,938 735,936 731,939 720,937 716,940 714,945 715,943 713,947 713,945 716,944 712,946 710,944 706,947 700,945 696,948 682,946 678,948 671,947 667,949 646,948 648,948 649,949 646,944 636,943 632,945 635,944 631,946 631,944 627,947 608,945 604,948 599,946 595,948 597,947 592,950 588,948 584,951 569,949 565,952 550,950 552,951 548,949 550,950 498,948 494,951 496,949 492,952 480,950 476,952 476,951 472,953 468,952 471,952 466,951 462,953 461,952 457,954 450,952 452,953 448,952 450,952 444,951 446,952 439,950 440,950 442,952 439,947 430,945 425,951 426,948 416,947 412,949 411,948 407,950 402,948 398,951 365,949 361,952 304,950 307,951 295,949 291,952 293,950 288,957 289,955 292,953 287,959 288,956 286,969 288,973 287,972 289,976 288,978 290,982 294,984 292,983 313,984 311,984 336,985 340,983 338,984 345,979 379,980 383,978 389,980 387,979 394,980 398,978 406,980 404,979 408,980 406,980 411,982 416,979 416,980 420,978 467,980 471,977 524,979 528,976 529,978 527,977 542,979 546,976 604,978 603,978 601,976 608,980 605,980 616,981 620,979 621,975 621,976 619,978 623,976 632,977 636,975 653,976 657,974 660,976 664,973 670,975 674,972 680,974 684,972 690,973 694,971 699,972 703,970 704,972 708,969 709,971 713,968 711,970 715,968 712,969 717,966 717,968 721,965 724,967 728,964 728,966 732,964 732,965 736,963 736,964 740,962 739,964 743,961 742,963 746,960 745,962 749,960 748,961 752,959 749,960 753,958 752,960 756,957 758,953 757,956 759,940 756,936 748,935"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0004_line_0002">
|
||||
<pc:Coords points="731,963 725,967 728,965 724,968 724,966 720,968 720,967 716,969 713,968 709,970 706,968 702,971 696,969 692,972 689,970 685,972 683,971 679,973 677,972 673,974 674,972 670,975 668,973 664,976 650,974 646,976 640,975 636,977 627,976 623,978 614,976 609,982 607,983 607,983 608,982 609,984 609,984 609,984 610,985 612,981 532,976 533,976 526,975 522,977 520,981 521,979 522,977 518,980 488,978 484,980 482,984 482,983 484,981 480,984 476,982 479,983 475,981 471,984 465,982 468,983 439,981 441,982 426,980 422,983 419,981 421,982 417,980 419,980 420,983 401,978 397,980 397,979 393,981 394,980 389,983 366,981 362,984 363,982 359,984 344,983 340,985 342,984 344,984 346,987 339,982 341,983 333,981 329,984 320,982 323,983 294,981 296,982 290,980 292,981 288,980 290,980 285,979 280,984 281,982 279,980 275,983 270,981 266,984 263,982 259,984 258,983 260,984 253,984 254,983 250,985 252,984 248,986 246,1008 273,1012 271,1012 281,1013 279,1012 289,1014 287,1013 292,1015 290,1014 304,1016 304,1016 304,1016 307,1010 305,1012 305,1012 305,1012 304,1013 314,1015 318,1012 319,1014 323,1012 324,1013 328,1011 330,1012 334,1010 341,1012 346,1006 345,1008 370,1010 368,1010 367,1008 369,1012 380,1013 384,1011 408,1012 405,1012 418,1013 422,1011 449,1012 454,1007 453,1009 455,1005 455,1007 462,1008 466,1006 545,1008 543,1007 541,1004 545,1010 552,1012 556,1008 558,1004 558,1006 556,1008 560,1005 626,1007 625,1008 633,1009 637,1007 641,1008 645,1006 664,1008 668,1005 685,1007 689,1004 694,1006 698,1004 696,1005 700,1003 701,998 702,999 702,1000 706,998 708,1000 712,997 714,999 718,996 717,998 722,995 720,996 724,994 722,996 726,993 724,995 728,992 730,994 734,992 734,993 738,991 739,992 743,990 742,992 746,989 745,991 749,988 747,990 752,987 753,983 752,985 747,986 747,986 751,989 756,986 757,982 756,984 758,978 757,980 759,968 741,964 744,964 731,963"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0004_line_0003">
|
||||
<pc:Coords points="466,1006 462,1008 452,1007 448,1009 430,1008 426,1010 407,1008 403,1011 392,1009 394,1010 380,1008 382,1009 357,1008 360,1008 346,1007 342,1009 334,1008 330,1010 328,1008 324,1011 324,1009 320,1012 318,1010 314,1012 312,1011 308,1013 306,1012 302,1014 292,1012 294,1013 286,1012 288,1012 278,1011 280,1012 266,1010 268,1011 250,1009 245,1036 248,1040 246,1038 264,1043 261,1042 284,1044 288,1041 299,1043 296,1042 337,1044 342,1040 341,1042 341,1042 345,1036 344,1038 344,1038 344,1038 344,1040 354,1042 358,1040 360,1036 359,1038 356,1040 360,1037 361,1039 365,1036 451,1038 448,1037 447,1035 450,1040 461,1041 466,1038 468,1033 468,1035 496,1036 500,1034 572,1036 576,1033 571,1034 591,1036 595,1033 596,1035 600,1032 597,1034 601,1032 603,1027 600,1023 602,1025 596,1020 599,1021 592,1020 594,1020 596,1016 593,1012 595,1015 528,1010 524,1012 527,1011 529,1012 522,1010 524,1011 526,1012 524,1008 525,1011 504,1006 500,1008 484,1007 487,1008 466,1006"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0005" orientation="1.241379310344827" type="paragraph">
|
||||
<pc:Coords points="736,664 731,664 728,664 724,664 724,664 719,666 719,664 716,668 716,668 716,666 712,668 656,668 652,668 620,668 618,673 618,671 604,671 606,671 606,671 582,668 584,668 575,668 572,668 467,668 464,673 464,671 450,671 452,671 452,671 419,668 416,673 416,673 414,676 414,676 414,676 416,676 416,676 416,676 419,673 419,673 419,673 412,668 414,668 409,668 407,668 400,668 397,673 397,671 371,671 366,671 335,671 332,673 304,671 301,676 301,676 299,673 301,673 301,676 299,671 294,676 294,676 294,673 289,676 292,676 289,695 289,695 287,700 287,700 287,700 287,700 287,700 284,702 284,702 284,702 277,700 277,700 277,700 277,700 260,700 260,700 277,702 277,702 277,702 292,695 292,695 289,697 289,697 289,678 289,678 292,680 289,676 282,676 284,676 265,676 268,676 253,673 251,676 251,676 248,676 248,683 248,680 246,697 246,695 246,702 246,702 248,700 244,719 248,721 246,721 246,786 248,788 246,817 248,822 248,836 248,839 248,851 253,853 251,853 251,889 251,894 251,904 253,906 251,906 306,908 304,908 318,908 320,908 335,908 337,904 337,906 356,906 359,904 359,904 368,906 371,901 371,904 380,904 383,901 380,904 385,899 385,901 385,892 383,887 385,887 385,889 385,880 385,880 385,877 385,877 385,877 383,880 388,877 421,877 419,877 428,880 431,877 467,877 472,877 472,877 474,875 474,877 476,872 476,875 546,875 546,875 544,872 599,877 596,877 613,877 618,872 618,875 623,875 620,875 620,875 642,877 647,875 644,877 649,872 649,875 690,875 692,872 692,875 695,872 709,872 712,872 712,872 714,868 714,870 721,870 726,870 724,870 726,868 726,870 728,868 726,868 731,865 731,865 750,868 752,865 752,865 757,858 757,860 757,856 757,858 760,856 760,856 757,858 760,750 757,748 757,748 760,743 760,745 757,745 760,666 757,664 757,664" conf="0.750609157462953"/>
|
||||
<pc:TextLine id="region_0005_line_0001">
|
||||
<pc:Coords points="734,660 730,663 730,661 726,664 726,662 721,665 720,671 720,671 720,671 721,673 722,671 724,669 720,672 716,670 712,672 707,671 703,673 698,672 694,674 670,672 666,675 664,673 666,674 660,672 662,673 664,675 660,670 655,668 651,671 652,669 648,672 650,670 646,672 647,671 649,672 635,670 631,672 619,671 621,672 614,670 610,672 609,671 612,672 604,670 607,671 603,669 605,670 533,668 536,669 523,668 525,668 513,667 509,669 492,668 495,668 476,667 472,669 467,668 463,670 456,668 452,671 444,669 440,672 418,670 418,670 418,670 415,673 417,674 417,674 417,674 420,674 416,668 411,667 413,668 409,666 405,668 400,667 395,672 396,670 370,668 366,671 343,669 339,672 344,671 336,669 332,672 304,670 304,670 300,674 300,672 302,673 302,673 298,670 293,676 294,674 293,672 288,692 291,696 289,696 295,701 292,700 308,702 312,700 315,701 319,699 336,700 340,698 363,700 367,697 420,699 418,698 426,700 424,700 423,697 425,701 437,703 442,700 475,701 472,700 488,702 493,698 498,700 502,697 683,699 687,696 692,698 696,696 708,697 712,695 718,696 722,694 725,696 729,693 734,695 738,692 736,694 740,692 738,693 742,691 742,692 746,690 748,686 752,683 750,684 748,684 752,685 756,683 757,679 756,681 758,676 757,678 759,668 756,664 747,662 749,663 745,661 748,662 734,660"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0005_line_0002">
|
||||
<pc:Coords points="740,688 736,690 737,688 732,692 735,690 731,692 730,691 726,693 725,692 721,694 722,692 718,695 718,693 714,696 700,694 696,696 693,695 696,696 687,694 682,697 683,696 679,698 674,696 670,699 660,697 656,700 640,698 636,700 628,699 631,700 626,698 628,699 622,697 618,700 600,698 596,700 591,699 587,701 576,700 579,700 574,699 576,699 544,697 540,700 534,698 529,701 528,706 528,704 431,703 433,704 435,704 432,700 396,698 398,699 393,697 389,700 383,698 379,700 303,699 299,701 289,700 285,702 286,700 282,703 284,701 286,702 276,700 272,703 269,701 270,700 266,699 268,699 260,697 256,700 255,704 256,701 254,705 254,704 253,702 249,704 251,703 247,705 245,709 246,707 244,726 287,731 291,728 329,730 333,728 343,729 347,727 370,728 368,728 408,729 405,728 409,730 407,729 405,727 408,732 418,733 422,731 427,732 431,730 430,732 435,726 434,728 432,730 436,728 443,729 447,727 512,728 509,728 513,729 511,728 509,726 512,730 533,732 532,732 550,733 554,731 561,732 565,730 569,732 567,731 576,728 574,730 572,729 572,729 584,732 589,725 590,726 588,728 585,727 592,726 652,728 649,727 648,724 652,730 659,732 663,729 680,731 684,728 685,730 689,728 704,729 708,727 709,728 707,728 714,727 716,728 720,726 717,728 721,725 723,720 722,723 724,719 723,721 720,723 724,720 727,722 731,720 732,721 736,719 735,720 739,718 740,720 744,717 744,719 748,716 749,712 750,713 752,715 756,712 753,714 758,693 752,688 755,689 740,688"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0005_line_0003">
|
||||
<pc:Coords points="740,715 736,718 738,716 733,720 733,718 729,720 730,719 726,721 728,720 724,722 727,720 723,723 725,721 721,724 723,722 719,724 720,723 716,725 708,724 703,731 703,731 704,728 704,728 704,728 704,728 705,732 705,732 705,732 705,732 689,724 685,727 660,725 656,728 637,726 632,732 632,732 632,732 632,732 635,736 636,733 636,732 636,732 632,733 577,732 580,732 581,734 578,729 564,728 560,730 530,728 532,729 520,728 522,728 513,727 516,728 496,726 498,727 490,725 486,728 488,726 484,728 481,727 477,729 443,728 445,728 431,727 427,729 387,728 383,730 381,735 382,732 380,736 381,734 383,732 379,735 377,733 380,734 366,732 362,735 325,733 327,733 328,734 326,730 305,728 308,729 298,728 294,730 274,728 270,731 250,729 246,732 244,758 247,762 245,761 259,766 263,764 281,765 285,763 283,764 287,762 287,764 291,761 298,763 302,760 305,762 303,761 316,763 320,760 324,762 328,760 336,761 336,761 335,760 333,759 350,764 354,761 362,763 360,762 376,764 380,761 382,756 382,758 425,760 423,759 427,760 424,760 431,761 428,760 439,762 443,760 443,761 447,759 478,760 476,760 484,761 488,759 485,760 483,760 491,761 495,759 502,760 506,758 504,760 502,759 509,758 512,760 516,757 520,759 524,756 551,758 555,756 613,757 612,758 619,760 623,757 623,759 627,756 624,758 628,756 632,757 636,755 656,756 653,756 664,757 662,756 684,758 689,752 690,753 696,755 700,752 704,754 708,752 707,753 712,750 714,752 718,749 722,751 726,748 727,750 731,748 732,749 736,747 740,748 744,746 744,748 748,745 745,747 750,740 749,743 751,738 752,739 749,740 753,738 752,740 756,737 758,733 757,736 759,724 756,720 746,718 748,719 750,720 748,716 740,715"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0005_line_0004">
|
||||
<pc:Coords points="744,747 740,749 729,748 725,750 720,748 716,751 717,749 713,752 712,750 708,752 709,751 705,753 706,752 702,754 704,752 698,757 700,756 696,758 697,756 693,759 692,757 688,760 672,758 668,760 637,759 633,761 634,760 636,760 632,759 634,760 636,760 629,755 632,756 628,754 624,756 629,756 624,754 620,756 619,755 615,757 596,756 598,756 584,755 580,758 578,763 578,761 520,760 520,759 510,757 512,758 508,756 504,759 482,757 477,763 478,760 476,764 476,763 454,761 450,764 406,762 408,763 410,763 404,758 407,759 403,757 399,760 401,758 397,760 396,765 396,763 398,761 394,764 394,762 396,763 391,761 387,764 354,762 350,764 351,763 353,764 347,762 349,763 328,761 330,762 323,760 324,760 288,759 284,761 287,760 281,764 268,762 264,764 253,763 256,764 251,762 247,764 249,763 244,785 247,789 254,791 258,788 286,790 284,789 288,791 285,790 295,792 299,789 302,791 306,788 316,790 320,788 331,789 335,787 380,788 377,788 381,789 379,788 377,787 380,792 386,793 390,791 390,792 394,790 408,792 406,791 411,792 415,790 418,792 422,789 426,791 430,788 437,790 435,789 439,791 436,790 443,792 448,786 447,788 444,790 449,784 450,785 452,787 449,786 456,785 483,787 487,784 620,786 617,785 622,787 620,786 624,788 622,787 628,788 635,784 644,785 648,783 670,784 674,782 696,784 700,781 716,783 713,782 720,781 718,783 722,780 720,782 728,776 727,778 731,776 728,777 732,775 731,776 735,774 734,776 738,773 736,775 740,772 738,774 742,772 740,773 744,771 747,772 751,770 748,772 752,769 752,771 756,768 758,760 752,755 754,756 750,754 752,754 753,754 751,750 752,752 750,748 744,747"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0005_line_0005">
|
||||
<pc:Coords points="751,770 747,772 748,771 744,773 742,772 738,774 736,772 732,775 732,773 728,776 730,774 726,776 728,775 724,777 726,776 722,778 724,776 720,779 721,777 717,780 717,778 713,780 709,779 705,781 685,780 685,780 685,780 685,780 685,780 685,780 691,790 691,790 684,786 684,786 688,788 688,788 688,788 681,780 684,780 679,779 675,781 676,780 672,782 674,780 670,783 668,781 663,784 661,788 661,787 632,785 628,788 625,786 628,787 621,785 617,788 623,787 616,788 616,786 618,787 609,785 612,786 600,784 603,785 599,784 595,786 566,784 562,787 564,785 560,788 548,786 544,788 530,787 532,788 507,786 509,787 496,785 492,788 492,788 491,790 493,788 496,789 481,788 477,790 444,788 440,791 443,789 439,792 439,790 441,791 443,792 438,786 430,784 426,787 420,785 416,788 391,786 386,789 387,788 383,790 380,788 380,788 376,792 376,791 378,792 359,790 355,792 342,791 344,792 346,792 344,788 345,791 317,786 313,788 252,787 248,789 250,788 245,813 248,817 256,819 253,818 282,820 280,819 284,820 282,820 297,822 301,820 325,821 329,819 329,820 327,820 337,821 341,819 340,820 344,818 345,814 345,816 354,817 358,815 392,816 389,816 396,817 394,817 399,819 403,816 401,818 405,816 406,817 410,815 432,816 436,814 478,816 482,813 561,815 559,814 568,816 565,815 569,816 567,816 572,817 569,816 573,818 577,816 576,817 580,815 577,816 582,813 584,815 588,812 640,814 638,813 644,815 642,815 648,816 645,816 655,817 659,815 671,816 675,814 678,816 682,813 701,815 705,812 707,814 711,812 709,813 713,811 713,812 717,810 716,812 720,809 719,811 723,808 725,810 729,808 731,809 735,807 735,808 739,806 736,808 741,804 739,806 744,803 745,799 744,801 746,797 747,798 744,800 748,797 747,799 751,796 752,798 756,796 758,788 757,791 759,777 756,773 758,775 751,770"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0005_line_0006">
|
||||
<pc:Coords points="748,797 744,800 746,798 742,800 744,799 739,803 741,801 737,804 739,802 735,804 735,803 731,805 731,804 727,806 723,804 719,807 720,805 716,808 719,806 715,808 715,807 711,809 711,808 707,810 703,808 699,811 684,809 680,812 676,810 672,812 664,811 660,813 647,812 649,812 641,811 644,812 639,810 634,813 629,812 625,814 624,819 624,816 623,820 624,818 611,816 607,819 574,817 576,817 577,818 571,813 573,814 556,812 552,815 510,813 506,816 502,814 504,815 490,813 492,814 482,812 478,815 474,813 470,816 468,820 469,817 469,817 468,821 470,820 470,820 433,819 436,820 432,818 428,820 417,819 420,820 414,818 416,819 408,817 410,817 412,820 390,815 392,816 384,814 380,816 365,815 368,816 352,814 348,816 342,815 338,817 320,816 316,818 302,816 298,819 300,817 296,820 264,818 267,819 252,817 247,835 249,839 248,841 255,846 252,845 271,847 275,844 328,846 332,844 359,845 363,843 381,844 379,844 383,845 387,843 392,844 390,844 420,845 420,845 421,844 420,845 419,843 422,848 430,849 435,846 436,842 436,844 436,846 440,844 450,845 454,843 452,844 450,844 454,845 458,843 472,844 470,844 516,845 520,843 520,844 517,844 516,841 519,846 528,848 532,845 533,847 537,844 543,846 547,844 556,845 554,844 586,846 590,844 601,845 599,844 615,846 619,844 624,845 621,844 644,846 648,844 651,845 648,844 653,846 657,844 668,845 672,843 671,844 675,842 676,844 680,841 684,843 688,840 700,842 704,840 706,841 710,839 712,840 716,838 717,840 721,837 724,839 728,836 726,838 732,834 729,836 733,833 731,835 736,832 735,833 739,831 742,832 746,830 746,832 750,829 748,831 752,828 751,830 756,827 757,823 756,825 758,818 757,820 759,814 756,810 758,803 752,798 755,799 748,797"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0005_line_0007">
|
||||
<pc:Coords points="745,826 741,828 744,827 740,829 741,828 737,830 739,828 735,831 737,829 733,832 733,832 733,832 733,832 734,834 736,834 736,834 732,831 728,833 730,832 726,834 727,832 723,835 724,833 720,836 718,834 714,836 711,835 707,837 703,836 699,838 691,836 693,837 686,838 687,836 683,839 682,837 678,840 679,838 675,840 677,839 672,845 673,843 672,848 672,845 671,844 667,846 625,844 628,845 621,844 623,844 624,846 622,842 592,840 588,843 556,841 559,842 546,840 542,843 534,841 530,844 532,842 528,844 491,843 486,850 487,848 484,846 480,848 468,847 470,848 443,846 439,848 436,847 439,848 440,849 438,845 424,844 427,844 404,843 400,845 399,844 394,847 363,845 364,845 366,848 364,844 360,842 362,843 358,841 360,842 354,840 349,844 352,842 348,844 340,843 336,845 339,844 334,849 335,847 333,852 333,850 275,848 276,848 278,851 276,847 272,845 274,846 254,844 250,847 248,868 251,872 249,870 256,875 254,874 258,876 256,875 260,876 257,876 256,873 258,877 256,875 264,880 268,877 265,879 269,876 267,878 272,875 272,876 276,874 273,876 277,873 317,875 321,872 319,874 316,873 322,875 326,872 370,874 368,873 372,875 370,875 368,872 371,876 379,878 383,876 383,877 387,875 387,876 391,874 415,876 412,875 420,876 418,876 424,877 421,876 435,878 439,876 452,877 456,875 465,876 469,874 471,870 471,872 469,873 473,871 506,872 510,870 604,872 601,871 607,872 604,872 608,873 606,872 610,874 608,873 620,875 624,872 624,874 628,872 650,873 648,872 655,872 649,872 653,874 651,873 664,875 668,872 677,874 681,872 681,873 685,871 684,872 688,870 688,872 692,869 691,871 695,868 702,870 706,868 707,869 704,868 712,870 716,868 716,869 720,867 717,868 722,863 723,864 720,865 724,863 735,864 739,862 741,864 745,861 744,863 748,860 746,862 751,856 750,859 748,860 756,855 757,850 756,852 758,848 757,850 759,835 756,831 758,833 752,828 755,829 751,828 752,828 745,826"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0005_line_0008">
|
||||
<pc:Coords points="309,872 305,875 308,873 301,878 280,876 276,879 278,877 274,880 256,878 252,880 253,879 248,890 251,894 249,898 252,902 294,904 292,903 305,904 304,904 302,902 304,906 303,904 320,908 324,906 327,908 331,905 329,907 334,900 333,902 331,904 335,901 348,903 352,900 374,902 378,900 377,901 382,895 381,897 383,893 380,889 382,892 380,888 372,886 375,887 376,889 374,885 376,883 372,878 351,876 353,877 349,876 352,876 353,879 350,874 309,872"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0006" orientation="1.241379310344827" type="paragraph">
|
||||
<pc:Coords points="656,203 652,203 630,203 625,205 620,203 620,203 620,203 618,212 618,212 618,212 616,203 611,208 611,208 572,205 568,208 553,208 553,208 505,205 503,208 467,208 462,208 455,208 450,210 438,208 436,212 436,210 436,210 438,210 438,212 431,208 428,212 428,210 400,210 402,210 402,212 395,208 392,212 392,210 366,210 364,215 364,212 347,212 349,212 349,212 330,210 328,215 328,212 325,212 328,212 328,212 292,210 289,215 289,212 289,212 287,217 287,215 287,227 287,229 287,239 287,236 287,241 287,239 287,239 282,239 284,239 280,241 280,239 277,241 275,241 275,241 272,239 268,241 268,241 270,241 263,239 260,241 253,241 251,244 251,244 251,241 248,248 248,246 248,306 248,304 246,349 248,352 248,407 248,409 248,409 248,409 248,464 248,467 248,464 253,469 251,469 251,467 251,469 263,472 260,472 260,469 301,472 304,469 304,469 344,472 349,469 371,469 373,467 373,467 388,469 388,469 385,467 407,469 412,467 412,467 414,469 414,469 412,467 433,469 438,467 493,469 493,469 491,467 503,469 508,467 508,467 510,469 508,469 508,467 522,469 524,467 524,467 532,469 534,464 534,467 534,440 534,440 534,443 536,438 536,440 534,440 539,436 539,436 606,438 604,438 620,438 618,438 630,438 628,438 628,438 642,440 642,440 642,440 644,438 642,438 642,438 647,440 649,436 649,438 652,438 654,438 659,438 661,436 673,438 678,436 678,436 680,433 680,433 690,436 692,433 709,433 712,433 714,433 716,431 736,433 738,428 738,431 748,431 750,428 750,431 755,428 752,428 757,428 755,428 757,424 757,426 757,426 760,424 762,308 762,311 762,272 762,272 762,248 762,246 762,239 762,241 762,234 762,232 762,232 762,234 762,229 762,227 762,224 762,227 762,217 762,215 762,208 760,203 750,203 752,203 695,203 692,203 690,203 692,203 678,203 676,203 656,203 659,203" conf="0.780629329792061"/>
|
||||
<pc:TextLine id="region_0006_line_0001">
|
||||
<pc:Coords points="678,200 674,203 668,201 664,204 664,202 660,204 658,203 658,203 658,203 657,210 657,210 658,206 650,204 646,207 620,205 620,205 620,205 618,211 618,211 618,211 612,205 615,206 572,204 568,207 552,205 555,206 545,204 541,207 527,205 529,206 505,204 501,207 489,205 485,208 488,206 484,208 474,207 470,209 439,208 435,210 436,208 438,209 434,208 430,210 400,208 403,209 404,212 396,207 391,212 392,210 366,208 361,214 362,212 347,210 349,211 351,213 330,208 330,208 326,212 326,210 328,211 328,211 292,208 287,214 288,212 290,210 285,216 286,214 284,227 287,231 285,235 291,240 288,239 293,240 291,240 296,241 294,240 301,242 299,241 308,243 312,240 336,242 340,240 361,241 365,239 407,240 411,238 415,240 419,237 442,239 440,238 452,240 450,239 480,240 484,238 491,240 488,239 496,240 493,240 497,241 495,240 500,242 498,241 506,243 510,240 509,242 513,240 512,241 516,239 516,240 520,238 520,240 524,237 542,239 540,238 560,240 557,239 575,240 579,238 582,240 586,237 596,239 594,238 600,240 598,239 618,240 622,238 638,240 636,239 649,240 653,238 653,240 657,237 672,239 676,236 687,238 691,236 701,237 705,235 704,236 702,236 713,237 717,235 720,236 724,234 725,236 729,233 730,235 734,232 737,234 741,232 743,233 747,231 752,232 756,230 759,232 764,224 763,227 764,217 762,213 764,207 761,203 751,201 753,202 728,200 725,209 724,210 724,210 724,210 724,210 724,210 724,210 723,210 723,210 723,210 723,210 723,210 723,210 728,204 728,204 720,201 716,204 711,202 706,208 706,208 706,208 706,208 711,212 712,209 712,209 710,205 693,204 693,204 693,204 693,204 693,204 692,208 692,208 692,208 694,208 694,208 692,203 688,201 690,202 678,200"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0006_line_0002">
|
||||
<pc:Coords points="751,228 747,230 744,228 740,231 735,229 731,232 732,230 728,232 728,231 724,233 725,232 721,234 696,232 692,235 680,233 676,236 657,234 653,236 654,235 650,237 650,236 646,238 637,236 640,237 622,236 618,238 600,236 602,237 596,236 598,236 591,235 593,236 588,234 584,236 579,235 575,237 573,236 569,238 562,236 564,237 536,236 538,236 525,235 521,237 516,236 512,238 513,236 509,239 509,237 505,240 500,238 503,239 496,237 498,238 492,236 495,237 480,236 476,238 452,236 455,237 449,236 449,236 449,236 447,240 447,240 447,240 440,236 442,236 415,235 411,237 408,236 404,238 348,236 344,239 336,237 332,240 320,238 323,239 316,237 312,240 306,238 302,240 304,239 307,240 296,238 298,239 292,237 295,238 286,236 282,239 284,237 280,240 280,238 276,240 274,239 276,240 272,238 268,240 268,239 270,240 263,238 259,240 256,239 252,241 255,240 251,242 252,240 247,248 248,246 246,270 252,275 256,272 263,274 267,272 269,273 273,271 273,272 277,270 279,272 283,269 295,271 299,268 300,270 297,269 381,271 385,268 404,270 402,269 410,271 408,270 414,272 412,271 432,272 436,270 437,272 441,269 459,271 463,268 468,270 472,268 477,269 481,267 489,268 493,266 516,268 520,265 532,267 536,264 564,266 561,265 570,267 568,266 574,268 572,267 579,268 576,268 585,269 583,268 591,270 588,269 599,271 596,270 607,272 611,269 610,271 614,268 612,270 616,268 615,269 619,267 616,268 620,266 620,268 624,265 632,267 636,264 641,266 639,265 649,267 647,266 654,268 652,267 659,268 656,268 667,269 671,267 678,268 682,266 688,268 692,265 693,267 697,264 699,266 703,264 704,265 708,263 711,264 715,262 728,264 732,261 734,263 738,260 739,262 743,260 759,261 763,259 764,249 762,245 764,240 763,242 764,235 762,231 764,232 751,228"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0006_line_0003">
|
||||
<pc:Coords points="740,257 736,260 733,258 729,260 724,259 720,261 709,260 705,262 706,260 702,263 700,261 696,264 693,262 689,264 689,263 685,265 680,264 676,266 666,264 662,267 659,265 661,266 651,264 653,265 645,264 648,264 639,263 641,264 632,262 628,264 622,263 618,265 620,264 616,266 618,264 614,267 616,265 612,268 613,266 609,268 592,267 595,268 584,266 587,267 578,265 580,266 572,264 574,265 568,264 571,264 565,263 568,264 525,262 521,264 512,263 508,265 510,264 512,264 498,263 494,265 490,264 486,266 477,264 473,267 467,265 463,268 456,266 452,268 452,267 454,268 440,266 436,268 436,267 432,269 411,268 413,268 404,267 407,268 398,266 400,267 389,265 385,268 382,266 378,268 314,267 316,268 282,266 278,268 279,267 275,269 275,268 271,270 270,268 266,271 264,269 260,272 255,270 251,272 251,271 246,294 270,299 268,298 275,300 272,299 277,300 275,300 280,301 277,300 285,302 289,300 308,301 306,300 316,302 314,301 337,303 341,300 362,302 366,300 368,301 372,299 371,300 375,298 372,300 376,297 374,299 378,296 377,298 381,296 381,297 385,295 412,296 409,296 422,297 420,296 428,298 426,297 435,299 432,298 444,300 448,297 448,299 452,296 453,298 457,296 464,297 461,296 468,298 466,297 474,299 472,298 478,300 476,299 486,300 484,300 495,301 499,299 500,300 504,298 504,300 508,297 510,299 514,296 517,298 515,297 523,299 520,298 526,300 530,297 532,299 536,296 536,298 540,296 543,297 547,295 547,296 544,296 554,297 552,296 561,298 559,297 563,299 567,296 571,298 575,296 578,297 582,295 586,296 590,294 592,296 596,293 597,295 601,292 604,294 608,292 614,293 612,292 625,294 623,293 632,295 629,294 656,296 660,293 670,295 668,294 673,296 671,295 676,296 674,296 679,297 676,296 684,298 682,297 688,299 692,296 704,298 708,296 711,297 715,295 714,296 718,294 717,296 721,293 720,295 724,292 724,294 728,292 726,293 730,291 730,292 734,290 746,292 750,289 756,291 760,288 758,290 763,284 762,287 764,272 763,274 764,268 762,264 764,264 761,260 763,263 756,258 759,259 740,257"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0006_line_0004">
|
||||
<pc:Coords points="755,286 751,288 740,287 736,289 732,288 728,290 728,288 724,291 727,289 723,292 724,290 720,292 720,291 716,293 717,292 713,294 714,292 710,295 702,293 698,296 680,294 682,295 675,293 677,294 672,292 675,293 670,292 672,292 664,291 667,292 661,290 657,292 655,291 651,293 632,292 635,292 625,291 628,292 615,290 617,291 606,289 602,292 600,290 596,292 594,291 590,293 587,292 583,294 580,292 576,295 568,293 564,296 559,294 561,295 548,293 550,294 540,292 536,295 535,293 531,296 529,294 525,296 523,295 525,296 508,294 504,296 506,295 502,297 501,296 497,298 479,296 481,297 472,296 474,296 468,295 470,296 464,294 466,295 452,293 448,296 450,294 446,296 448,295 444,297 433,296 436,296 428,295 431,296 422,294 424,295 410,293 412,294 381,292 377,295 379,293 375,296 377,294 372,297 375,296 371,298 372,296 368,299 364,297 360,300 357,298 360,299 344,297 340,300 342,298 344,299 337,300 327,298 323,300 321,299 324,300 312,298 314,299 305,297 308,298 276,296 279,297 272,296 275,296 268,295 270,296 255,294 251,296 252,295 248,298 246,306 247,304 245,326 276,331 280,328 284,330 288,328 292,329 296,327 304,328 308,326 306,328 304,327 332,328 336,326 340,328 344,325 346,327 350,324 366,326 364,325 383,327 380,326 395,328 399,325 434,327 438,324 445,326 443,325 451,327 448,326 457,328 455,327 464,328 464,328 464,328 464,328 467,321 465,323 465,323 465,323 468,328 472,326 476,328 480,325 483,327 487,324 487,326 491,324 490,325 494,323 495,324 499,322 516,324 514,323 596,324 593,324 612,325 609,324 617,326 615,325 636,327 640,324 642,326 646,324 664,325 668,323 675,324 672,324 683,325 683,325 683,325 686,322 684,321 684,321 684,321 684,324 688,326 686,325 700,327 704,324 704,326 708,324 708,325 712,323 712,324 716,322 716,324 720,321 720,323 724,320 726,322 730,320 731,321 735,319 734,320 738,318 736,320 740,317 738,319 742,316 740,318 744,316 743,317 747,315 746,316 750,314 751,316 755,313 756,315 760,312 758,314 763,308 762,311 764,292 761,288 755,286"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0006_line_0005">
|
||||
<pc:Coords points="754,310 750,312 751,311 747,313 748,312 744,314 747,312 743,315 744,313 740,316 743,314 739,316 740,315 736,317 736,316 732,318 732,316 728,319 728,317 724,320 724,318 720,320 719,319 715,321 715,320 711,322 712,320 708,323 708,321 704,324 700,322 696,324 690,323 692,324 681,322 684,323 672,321 674,322 664,320 660,323 648,321 644,324 637,322 633,324 621,323 624,324 609,322 612,323 597,321 600,322 584,320 587,321 556,320 552,322 524,320 527,321 506,320 508,320 501,319 497,321 497,320 493,322 494,320 490,323 491,321 487,324 484,322 480,324 475,323 471,325 452,324 455,324 447,323 449,324 432,322 428,324 418,323 420,324 402,322 398,324 372,323 375,324 347,322 343,324 340,323 336,325 328,324 324,326 319,324 321,325 298,324 294,326 292,324 288,327 284,325 280,328 272,326 268,328 251,327 247,329 245,349 248,353 246,351 268,356 265,355 278,356 276,356 289,357 293,355 303,356 307,354 312,356 316,353 323,355 327,352 357,354 355,353 363,355 360,354 367,356 364,355 370,356 368,356 372,357 370,356 382,358 386,356 384,357 388,355 386,356 390,354 389,356 393,353 394,355 398,352 406,354 404,353 432,355 429,354 474,356 478,353 482,355 486,352 489,354 493,352 494,353 498,351 502,352 506,350 512,352 516,349 528,351 526,350 550,352 554,349 585,351 589,348 607,350 604,349 616,351 613,350 628,352 632,349 637,351 641,348 656,350 653,349 674,351 672,350 683,352 687,349 697,351 701,348 704,350 708,348 711,349 715,347 720,348 724,346 728,348 732,345 734,347 738,344 746,346 750,344 756,345 760,343 758,344 763,316 757,311 760,312 754,310"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0006_line_0006">
|
||||
<pc:Coords points="744,341 740,344 732,342 728,344 727,343 723,345 720,344 716,346 712,344 708,347 707,345 703,348 698,346 694,348 688,347 684,349 668,348 671,348 656,347 659,348 642,346 638,348 632,347 628,349 613,348 616,348 604,347 607,348 588,346 584,348 573,347 576,348 566,346 562,348 556,347 552,349 528,348 531,348 512,347 508,349 504,348 500,350 495,348 491,351 492,349 488,352 489,350 489,350 485,353 485,353 485,353 485,353 487,356 487,356 487,356 487,356 482,350 478,352 473,351 469,353 464,352 466,352 458,351 454,353 421,352 424,352 400,351 403,352 399,350 395,352 392,351 388,353 389,352 385,354 388,352 384,355 384,353 380,356 373,354 376,355 368,353 371,354 366,352 368,353 362,352 364,352 356,351 358,352 345,350 348,351 330,349 326,352 317,350 313,352 309,351 305,353 301,352 297,354 269,352 272,353 260,352 263,352 251,351 246,380 265,384 263,384 292,385 289,384 298,386 296,385 318,387 322,384 328,386 332,384 336,385 333,384 345,386 349,384 354,385 358,383 363,384 367,382 374,384 378,381 376,383 374,382 388,384 386,383 405,384 403,384 422,385 420,384 440,386 444,384 452,385 456,383 456,384 460,382 459,384 463,381 464,383 468,380 474,382 478,380 492,381 490,380 495,382 492,381 500,383 497,382 522,384 526,381 528,383 532,380 533,382 537,380 590,381 588,380 593,382 591,381 599,383 596,382 600,384 604,381 606,383 610,380 610,382 614,380 615,381 619,379 630,380 634,378 640,380 644,377 648,379 652,376 656,378 653,377 661,379 659,378 665,380 663,379 670,380 668,380 680,381 677,380 686,382 684,381 697,383 701,380 722,382 726,380 728,381 732,379 732,380 736,378 735,380 739,377 737,379 741,376 740,378 744,376 743,377 747,375 746,376 750,374 752,376 756,373 756,375 760,372 758,374 763,347 760,343 744,341"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0006_line_0007">
|
||||
<pc:Coords points="752,371 748,373 749,372 745,374 746,372 742,375 743,373 739,376 739,374 735,376 735,375 731,377 731,376 727,378 725,376 721,379 717,377 713,380 709,378 712,379 707,377 703,380 694,378 690,380 687,379 689,380 683,378 685,379 674,377 676,378 664,376 666,377 660,376 662,376 656,375 659,376 648,374 644,376 641,375 637,377 631,376 627,378 616,376 612,379 612,377 608,380 606,378 602,380 598,379 600,380 592,378 594,379 584,377 586,378 578,376 574,379 534,377 530,380 532,378 528,380 525,379 521,381 498,380 500,380 494,379 496,380 491,378 493,379 485,377 488,378 482,376 478,379 474,377 470,380 468,378 464,380 461,379 457,381 458,380 454,382 452,380 448,383 410,381 412,382 388,380 391,381 375,380 377,380 370,379 366,381 364,380 360,382 356,380 352,383 322,381 318,384 296,382 298,383 294,381 290,384 292,382 295,383 264,381 266,382 251,380 246,407 248,411 247,410 296,415 300,412 304,414 308,412 350,413 348,412 356,414 354,413 369,415 373,412 372,414 376,412 376,413 380,411 380,412 384,410 412,412 409,411 428,412 432,410 445,412 449,409 464,411 468,408 476,410 474,409 502,411 500,410 519,412 523,409 536,411 540,408 541,410 545,408 544,409 548,407 559,408 556,408 560,409 558,408 564,410 561,409 568,411 566,410 572,412 569,411 576,412 573,412 584,413 588,411 589,412 593,410 592,412 596,409 595,411 599,408 598,410 602,408 604,409 608,407 636,408 634,408 641,409 639,408 644,410 648,408 653,409 657,407 666,408 664,408 688,409 692,407 696,408 700,406 707,408 704,407 718,408 722,406 736,408 740,405 740,407 744,404 742,406 746,404 745,405 749,403 749,404 753,402 755,404 759,401 757,403 761,400 763,376 760,372 752,371"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0006_line_0008">
|
||||
<pc:Coords points="760,400 752,401 751,400 747,402 748,400 744,403 744,401 740,404 739,402 735,404 726,403 722,405 696,404 692,406 692,404 688,407 655,405 657,406 650,407 656,406 648,407 639,405 641,406 634,404 636,405 627,404 623,406 604,404 600,407 600,405 596,408 597,406 593,408 595,407 591,409 590,408 586,410 572,408 574,409 565,408 568,408 561,407 564,408 559,406 561,407 556,405 559,406 554,404 556,405 549,404 545,406 548,404 544,407 544,405 540,408 539,406 535,408 520,407 516,409 498,408 500,408 476,407 478,408 459,406 455,408 436,407 432,409 428,408 424,410 410,408 412,409 379,408 375,410 376,408 372,411 373,409 369,412 371,410 367,412 357,411 360,412 351,410 353,411 337,409 340,410 309,408 305,411 303,409 299,412 295,410 291,412 251,411 246,438 260,443 264,440 270,442 274,440 279,441 276,440 283,442 280,441 284,443 282,442 286,444 284,443 289,444 287,444 312,445 316,443 315,444 319,442 317,444 321,441 320,443 324,440 325,442 329,440 339,441 343,439 356,440 354,440 364,441 362,440 368,442 365,441 372,443 369,442 409,444 407,443 425,444 429,442 442,444 446,441 475,443 479,440 491,442 495,440 528,441 525,440 534,442 538,440 540,436 540,437 632,439 630,438 634,440 632,439 636,440 633,440 640,441 644,439 642,440 646,438 648,440 652,437 650,439 654,436 660,438 664,436 676,437 680,435 684,436 688,434 700,436 704,433 713,435 717,432 726,434 730,432 734,433 738,431 739,432 743,430 743,432 747,429 746,431 750,428 751,430 755,428 754,429 759,426 757,428 761,425 763,404 757,400 760,400"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0006_line_0009">
|
||||
<pc:Coords points="344,437 337,438 329,436 325,439 323,437 319,440 320,438 316,440 319,439 315,441 315,440 311,442 287,440 289,441 284,440 286,440 281,439 284,440 279,438 281,439 266,437 262,440 261,438 257,440 251,439 246,460 252,465 249,464 260,466 257,465 278,467 277,468 276,468 278,472 291,474 296,471 297,464 296,467 298,463 297,465 295,467 299,464 339,466 343,464 357,465 361,463 415,464 412,464 416,465 414,464 412,463 415,467 413,465 427,470 432,466 434,462 434,462 433,460 432,463 451,464 455,462 461,464 459,463 465,464 469,462 475,464 472,463 481,464 485,462 493,464 492,464 490,464 492,468 499,470 503,468 519,469 523,467 520,468 525,463 524,465 526,461 525,464 527,465 531,463 528,464 533,459 532,461 534,457 534,459 536,455 535,457 536,446 534,442 536,443 529,438 532,439 485,437 481,440 470,438 466,440 436,439 432,441 398,440 400,440 396,439 392,441 375,440 377,440 368,439 371,440 365,438 368,439 359,437 361,438 349,436 352,437 343,436 339,438 344,437"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0007" orientation="1.241379310344827" type="header">
|
||||
<pc:Coords points="467,908 462,911 462,908 464,908 460,935 464,940 462,940 462,937 467,942 464,942 464,940 479,942 481,940 481,940 534,942 532,942 532,940 541,942 546,940 546,940 548,942 548,942 546,940 551,942 556,940 556,940 556,942 558,940 558,940 560,920 560,920 560,916 560,913 560,913 556,911 558,911 558,911 539,908 541,908 536,908 536,908 536,908 534,913 536,913 536,913 505,908 503,913 503,913 503,911 503,911 503,911 503,911 505,913 505,913 505,913 484,908 484,908"/>
|
||||
<pc:TextLine id="region_0007_line_0001">
|
||||
<pc:Coords points="470,905 466,908 467,906 462,909 460,916 461,913 460,917 462,921 460,924 463,928 461,933 464,937 462,936 476,940 480,938 480,940 484,937 512,939 509,938 548,940 552,937 552,939 556,936 558,931 557,933 559,917 556,913 552,912 555,912 539,911 541,912 537,910 537,910 533,914 535,912 537,913 539,915 496,910 498,911 480,909 483,910 479,908 480,908 482,911 476,906 479,907 470,905"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0008" orientation="1.241379310344827" type="header">
|
||||
<pc:Coords points="460,1060 457,1060 455,1060 455,1086 457,1091 457,1091 476,1093 481,1088 481,1091 534,1091 536,1088 548,1091 553,1088 556,1088 558,1088 558,1076 558,1079 558,1079 560,1067 556,1062 558,1062 558,1064 556,1060 556,1060 548,1060 546,1062 546,1060 548,1060 539,1060 536,1062 536,1060 536,1060 524,1060 524,1060 512,1060 515,1060 503,1057 500,1060 491,1060 491,1060 476,1057 479,1057 479,1060"/>
|
||||
<pc:TextLine id="region_0008_line_0001">
|
||||
<pc:Coords points="463,1055 459,1057 461,1056 456,1060 454,1075 456,1079 455,1083 457,1087 456,1085 458,1089 466,1091 470,1088 468,1090 465,1089 472,1091 476,1088 525,1090 529,1088 535,1089 539,1087 540,1088 544,1086 549,1088 555,1084 556,1080 557,1080 559,1075 556,1070 557,1071 555,1067 556,1072 557,1068 555,1064 556,1067 554,1063 550,1061 552,1062 473,1060 476,1061 472,1060 474,1060 476,1063 473,1059 475,1060 469,1056 472,1056 463,1055"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0009" orientation="1.241379310344827" type="header">
|
||||
<pc:Coords points="503,623 498,625 498,623 457,623 455,625 455,623 452,644 457,647 455,647 455,647 455,652 455,652 457,654 455,654 474,656 476,652 476,654 476,654 479,649 479,649 476,652 476,652 476,652 476,649 476,649 476,649 476,649 486,654 491,652 488,654 488,654 522,654 522,654 520,654 527,656 529,652 529,654 553,654 558,652 558,625 556,623 558,623"/>
|
||||
<pc:TextLine id="region_0009_line_0001">
|
||||
<pc:Coords points="459,621 455,624 457,622 452,628 453,626 452,633 454,637 452,640 455,644 453,645 456,649 454,647 461,652 459,651 464,652 468,650 486,652 490,649 492,651 490,650 494,652 498,649 498,651 496,650 500,652 497,651 502,652 500,652 514,653 518,651 529,652 533,650 533,652 537,649 535,651 539,648 536,650 542,646 541,648 545,645 550,647 554,644 556,630 553,626 555,628 549,624 552,624 530,623 532,624 528,622 524,624 484,623 487,624 472,622 474,623 459,621"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0010" orientation="1.241379310344827" type="header">
|
||||
<pc:Coords points="455,167 452,169 452,167 455,167 450,169 450,169 450,196 452,198 450,198 452,200 469,200 472,200 520,200 522,196 522,198 546,198 548,196 551,198 556,196 553,196 556,191 556,193 558,174 556,172 556,174 553,169 553,169 556,172 534,167 536,167 529,167 532,167 493,167 491,167 491,167 493,167 479,167 476,167 476,167 479,167 464,167 464,167 464,167 462,172 462,172 464,167"/>
|
||||
<pc:TextLine id="region_0010_line_0001">
|
||||
<pc:Coords points="460,164 456,167 458,165 454,168 455,166 451,168 449,172 450,170 448,193 451,197 449,196 455,200 452,200 472,201 476,199 480,200 485,197 484,199 488,196 491,198 488,197 494,199 494,199 494,199 497,196 495,195 495,195 495,195 492,195 496,200 494,197 519,202 524,196 523,198 537,200 541,197 540,199 544,196 544,198 548,196 548,197 552,195 551,196 555,194 552,196 556,193 558,187 557,189 559,177 556,173 558,176 556,172 557,174 555,170 550,168 552,169 548,168 550,168 536,167 538,168 526,166 522,168 513,167 509,169 500,168 496,170 475,168 477,169 472,168 474,168 470,167 472,168 468,166 470,167 464,165 464,165 464,165 462,169 462,169 466,169 460,164"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0011" orientation="1.241379310344827" type="header">
|
||||
<pc:Coords points="457,488 455,488 455,486 455,491 455,488 455,488 452,510 457,515 455,515 455,515 455,517 455,517 460,520 457,520 464,522 467,520 546,520 551,517 551,517 551,520 553,517 553,491 553,488 548,488 551,488 551,488 529,486 532,486 532,488 498,484 496,488 496,486 479,486 479,486 481,488"/>
|
||||
<pc:TextLine id="region_0011_line_0001">
|
||||
<pc:Coords points="460,484 456,487 459,485 454,488 452,504 455,508 453,513 456,517 460,519 457,518 464,520 468,517 475,519 479,516 484,518 488,516 495,517 499,515 498,516 496,516 500,517 497,516 504,518 501,517 508,519 505,518 511,520 515,517 521,519 525,516 531,518 535,516 536,517 540,515 537,516 541,514 539,516 544,512 544,513 548,511 548,512 553,500 552,503 554,495 548,490 551,491 533,489 536,490 529,488 525,491 511,489 507,492 502,490 504,491 484,489 486,490 476,488 479,489 473,488 475,488 471,486 472,486 460,484"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0012" orientation="1.241379310344827" type="header">
|
||||
<pc:Coords points="491,107 486,109 467,107 462,112 462,112 402,109 400,112 395,112 397,112 378,109 376,112 344,112 342,112 337,112 340,112 335,112 330,112 332,112 332,112 289,112 289,112 287,109 282,112 263,112 260,112 258,112 256,114 253,112 251,116 251,114 251,119 251,116 251,116 248,121 248,119 248,126 248,124 246,143 248,148 248,145 248,150 248,150 253,152 251,152 272,152 277,152 280,152 277,152 284,152 289,152 294,152 299,148 299,150 306,150 311,148 313,150 311,150 318,150 316,150 316,150 328,152 325,152 337,152 340,152 378,152 376,152 426,152 428,152 460,152 462,148 462,150 469,150 467,150 467,150 479,152 481,148 481,150 491,150 493,148 524,150 522,150 544,150 541,150 553,150 558,150 580,150 582,148 582,150 584,143 584,143 584,143 582,145 582,143 582,143 582,143 582,143 606,150 611,148 642,148 644,145 644,145 666,148 671,145 685,145 690,143 690,143 716,145 721,140 721,143 724,143 726,140 726,143 731,140 728,140 733,121 733,121 731,119 731,119 731,121 731,121 733,114 728,112 731,112 731,114 728,109 731,112 719,107 721,107 680,107 678,109 680,107 680,107 676,107 673,109 575,107 577,107 572,107 568,109 539,107 536,112 536,112 520,109 522,109 522,112 508,107 508,107"/>
|
||||
<pc:TextLine id="region_0012_line_0001">
|
||||
<pc:Coords points="689,105 685,108 603,106 599,108 601,107 597,109 599,108 595,110 565,108 561,111 536,109 539,110 520,108 522,109 517,108 513,110 513,108 509,111 512,109 508,112 496,110 492,112 483,111 485,112 472,110 474,111 464,109 466,110 403,108 399,111 395,109 397,110 383,108 379,111 362,109 358,112 356,110 359,111 345,109 341,112 338,110 340,111 334,109 330,112 332,110 334,111 321,109 317,112 320,110 316,112 317,111 313,113 316,112 311,115 300,113 303,114 289,112 292,113 283,112 285,112 274,111 276,112 266,110 262,112 258,111 254,113 254,112 250,114 248,118 249,116 252,114 247,120 248,118 246,125 247,123 245,144 248,148 246,146 252,151 249,150 253,152 251,151 256,152 253,152 261,153 259,152 264,154 261,153 269,155 273,152 288,154 292,152 295,153 300,148 299,150 307,152 311,149 312,151 310,150 318,152 316,151 314,148 323,153 320,152 330,154 328,153 337,155 341,152 348,154 352,152 350,153 354,151 396,152 393,152 415,153 419,151 420,152 424,150 424,152 428,149 432,151 436,148 485,150 489,148 532,149 530,148 556,150 556,150 556,150 558,146 558,146 558,146 555,147 579,152 583,149 581,151 581,151 581,151 581,151 582,145 582,145 582,145 582,145 582,145 607,151 611,148 640,150 640,150 640,150 640,150 639,140 638,143 638,143 640,147 667,148 671,146 686,148 691,142 690,144 711,146 715,144 717,145 721,143 724,144 728,142 727,144 732,140 729,142 734,122 734,122 731,118 732,120 732,122 732,122 734,116 731,111 732,113 730,109 726,108 728,108 722,107 724,108 719,106 721,107 689,105"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0013" orientation="1.241379310344827" type="marginalia">
|
||||
<pc:Coords points="209,221 209,222 208,223 133,223 133,276 134,276 135,276 135,278 134,279 133,279 133,293 134,293 135,294 135,318 134,319 133,319 133,336 134,336 135,336 135,337 136,337 137,338 137,339 151,339 151,338 152,337 174,337 174,336 175,336 198,336 199,335 219,335 220,334 221,334 222,333 226,333 226,331 227,330 227,328 228,328 228,326 228,325 229,325 229,324 231,323 231,322 232,321 232,322 242,322 243,321 243,320 244,319 244,318 244,317 245,317 245,314 246,313 246,304 245,304 245,302 246,301 246,292 247,292 247,280 248,280 248,268 248,267 249,267 249,256 248,256 248,256 248,240 247,240 247,221 209,221"/>
|
||||
<pc:TextLine id="region_0013_line_0001">
|
||||
<pc:Coords points="212,219 207,224 208,222 140,220 136,223 138,221 134,224 136,222 132,224 131,247 140,252 138,251 152,252 149,252 158,253 156,252 169,254 173,252 174,253 178,251 180,252 184,250 187,252 191,249 200,251 204,248 228,250 232,248 235,249 239,247 240,248 244,246 243,248 248,224 212,219"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0013_line_0002">
|
||||
<pc:Coords points="241,244 237,246 236,244 232,247 202,245 198,248 192,246 188,248 186,247 182,249 180,248 176,250 173,248 169,251 148,249 150,250 137,248 132,252 131,272 136,276 134,276 132,274 148,279 146,278 180,280 178,279 204,280 208,278 209,280 213,277 214,279 218,276 224,278 228,276 240,277 244,275 244,276 248,264 248,266 245,268 250,259 244,254 247,255 248,248 241,244"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0013_line_0003">
|
||||
<pc:Coords points="220,273 216,276 212,274 208,276 206,275 202,277 174,276 176,276 145,275 141,277 137,276 133,278 136,276 131,289 136,294 134,293 132,305 135,309 140,311 144,308 145,310 149,308 153,309 157,307 165,308 169,306 210,308 208,307 216,308 213,308 217,309 222,306 223,308 227,305 235,307 239,304 237,306 241,304 239,305 245,300 247,278 220,273"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0013_line_0004">
|
||||
<pc:Coords points="168,303 164,305 160,304 156,306 153,304 149,307 148,305 144,308 139,306 135,308 137,307 132,320 133,318 136,316 131,332 136,336 134,336 132,333 139,338 136,337 135,335 147,340 152,334 151,336 164,338 164,338 168,335 167,336 164,336 169,337 173,335 175,336 179,334 177,336 175,335 180,336 184,334 208,336 212,333 213,335 217,332 215,334 220,331 221,327 220,329 222,322 216,317 219,318 220,309 214,304 216,305 195,304 197,304 188,303 184,305 183,304 185,304 168,303"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0014" orientation="1.241379310344827" type="marginalia">
|
||||
<pc:Coords points="136,537 136,538 136,539 135,539 135,540 134,540 133,540 133,592 134,592 135,592 135,594 136,596 136,597 147,597 148,598 163,598 164,597 164,597 165,598 177,598 178,597 182,597 182,596 184,593 185,593 185,592 186,591 190,591 190,579 189,578 189,576 188,576 188,576 188,574 188,573 190,573 190,572 191,571 193,571 193,568 194,567 200,567 200,566 200,565 226,565 226,564 227,564 229,564 230,563 230,540 229,540 229,539 228,539 227,538 227,537 176,537 176,538 176,539 148,539 147,538 147,537 136,537"/>
|
||||
<pc:TextLine id="region_0014_line_0001">
|
||||
<pc:Coords points="179,535 174,540 175,538 157,536 152,540 138,539 134,541 136,540 132,545 132,543 131,551 133,555 132,559 134,563 132,561 138,566 136,565 148,567 152,564 164,566 168,564 194,565 198,563 199,564 203,562 197,563 204,562 203,564 200,563 210,564 214,562 212,564 216,558 216,560 216,561 220,559 224,560 228,558 229,554 228,556 230,548 228,544 223,542 225,543 217,541 219,541 215,540 217,540 213,539 215,539 216,541 213,536 179,535"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0014_line_0002">
|
||||
<pc:Coords points="141,564 137,567 138,565 133,572 134,570 132,586 135,590 140,592 137,591 136,588 144,593 141,592 165,594 163,593 175,595 180,592 181,588 180,590 182,580 178,575 179,580 174,572 176,573 172,572 175,572 176,575 173,570 163,568 165,569 160,568 156,570 158,568 160,569 154,568 156,568 158,571 156,567 152,565 154,566 141,564"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0015" orientation="1.241379310344827" type="marginalia">
|
||||
<pc:Coords points="200,684 199,684 177,684 177,686 176,687 165,687 164,688 149,688 148,688 136,688 135,689 135,691 134,692 133,692 133,714 134,714 135,715 135,719 134,720 133,720 133,803 134,803 135,804 136,804 137,804 137,806 162,806 162,804 163,804 177,804 180,801 193,801 193,800 195,799 202,799 203,798 204,798 204,796 204,796 205,796 207,794 207,790 208,789 208,787 208,786 209,786 210,785 210,784 211,784 211,783 212,782 212,779 212,778 213,779 213,784 214,784 215,784 215,785 216,786 220,786 220,787 220,788 227,788 228,788 228,789 235,789 235,788 236,788 236,786 236,785 237,785 237,784 238,784 238,780 239,779 239,772 240,771 240,768 240,768 241,768 241,747 242,746 242,723 243,722 243,718 244,717 244,689 243,688 243,684 242,684 229,684 228,684 224,684 224,684 200,684"/>
|
||||
<pc:TextLine id="region_0015_line_0001">
|
||||
<pc:Coords points="216,683 212,685 180,684 176,686 171,684 167,687 164,685 160,688 147,686 143,688 137,687 132,693 133,691 132,704 134,708 132,708 184,712 188,710 185,712 183,711 189,712 193,710 192,712 189,711 195,712 192,712 191,709 193,713 203,715 207,712 208,708 209,709 214,711 218,708 230,710 234,708 234,708 234,708 234,708 234,708 232,703 232,703 232,703 232,703 232,703 232,706 236,708 240,705 242,695 233,690 233,690 233,690 231,692 233,693 235,692 232,688 234,691 228,686 229,685 225,684 228,684 216,683"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0015_line_0002">
|
||||
<pc:Coords points="193,713 189,716 175,714 171,716 142,715 138,717 139,716 135,718 133,722 134,720 132,738 135,742 133,740 136,744 145,745 149,743 151,744 156,740 154,742 158,740 170,741 174,739 193,740 197,738 222,740 226,737 228,739 232,736 234,720 232,716 227,715 229,716 223,714 225,715 193,713"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0015_line_0003">
|
||||
<pc:Coords points="137,744 133,746 132,763 134,767 132,768 136,773 145,775 149,772 151,768 151,769 180,771 184,768 201,770 205,768 213,769 217,767 224,768 228,766 228,768 232,765 233,767 238,761 237,764 239,753 230,748 231,748 185,746 188,747 182,745 184,746 165,744 161,747 150,745 152,746 145,744 148,745 137,744"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0015_line_0004">
|
||||
<pc:Coords points="157,772 153,775 155,773 155,773 151,780 151,778 148,776 144,779 137,777 133,780 132,797 134,801 132,799 145,804 149,801 171,803 176,789 173,785 175,788 175,788 175,788 169,784 171,786 171,786 171,786 172,788 173,778 171,774 157,772"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0016" orientation="1.241379310344827" type="marginalia">
|
||||
<pc:Coords points="207,937 207,938 206,939 181,939 180,940 165,940 164,940 149,940 148,941 136,941 136,943 135,944 135,946 134,947 133,947 133,966 134,966 135,967 135,968 134,968 133,968 133,1082 136,1082 136,1083 156,1083 156,1082 190,1082 190,1080 191,1080 194,1080 195,1079 201,1079 202,1078 202,1068 201,1067 201,1065 200,1065 200,1064 200,1064 199,1063 199,1059 200,1058 204,1058 204,1059 208,1059 209,1060 208,1061 208,1063 206,1064 205,1064 204,1065 204,1066 204,1067 204,1077 206,1077 207,1078 208,1078 208,1079 225,1079 227,1077 227,1076 228,1076 228,1064 227,1064 227,1060 226,1060 226,1056 227,1056 227,1051 228,1050 229,1050 230,1049 230,1048 232,1046 233,1046 233,1036 234,1035 236,1035 236,1030 236,1029 237,1029 237,1027 239,1025 239,1024 240,1022 241,1022 241,1012 244,1010 244,1007 244,1006 245,1006 245,1002 246,1001 246,988 247,987 247,966 248,965 248,963 248,962 249,962 249,941 248,941 247,940 247,937 207,937"/>
|
||||
<pc:TextLine id="region_0016_line_0001">
|
||||
<pc:Coords points="212,935 208,937 209,936 205,938 184,936 180,939 168,937 164,940 152,938 148,940 140,939 136,941 138,940 133,945 134,943 132,948 133,946 136,944 131,960 133,964 132,962 134,966 132,964 145,969 149,967 174,968 178,966 222,968 220,967 228,968 232,966 242,968 247,964 248,959 248,961 245,963 250,945 248,941 244,940 245,940 241,938 244,939 240,937 242,938 237,936 239,936 212,935"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0016_line_0002">
|
||||
<pc:Coords points="199,964 195,966 170,964 166,967 147,965 143,968 137,966 133,968 132,972 132,970 131,990 133,994 132,993 134,997 151,999 155,996 158,998 162,996 166,997 170,995 174,996 178,994 184,996 188,993 202,995 200,994 220,996 224,993 237,995 241,992 239,994 243,992 244,988 244,990 245,986 244,988 246,975 244,971 245,971 242,966 209,964 212,965 199,964"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0016_line_0003">
|
||||
<pc:Coords points="186,992 182,994 176,992 172,995 168,993 164,996 160,994 156,996 152,995 148,997 138,996 134,998 136,996 132,999 131,1020 133,1024 132,1022 138,1027 136,1026 169,1028 173,1025 174,1027 178,1024 177,1026 181,1024 182,1025 186,1023 190,1024 194,1022 219,1024 224,1014 223,1016 224,1009 222,1005 224,1001 221,997 223,997 204,992 206,993 186,992"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0016_line_0004">
|
||||
<pc:Coords points="215,1020 211,1022 188,1020 184,1023 184,1021 180,1024 180,1022 176,1024 176,1023 172,1025 169,1024 165,1026 136,1024 132,1027 131,1050 136,1055 134,1054 154,1056 158,1053 165,1055 163,1054 172,1056 176,1053 185,1055 189,1052 188,1054 185,1053 201,1055 205,1052 207,1054 207,1054 207,1054 207,1054 207,1054 207,1047 207,1047 207,1051 216,1052 220,1050 221,1052 225,1049 224,1051 228,1048 225,1050 230,1042 229,1044 231,1032 228,1028 230,1029 228,1025 229,1026 226,1021 215,1020"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0016_line_0005">
|
||||
<pc:Coords points="152,1052 148,1054 136,1052 131,1077 133,1081 139,1083 136,1082 152,1084 156,1081 175,1083 179,1080 184,1082 188,1080 186,1081 190,1079 189,1080 193,1078 195,1073 194,1076 196,1057 193,1053 152,1052"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
<pc:TextRegion id="region_0017" orientation="1.241379310344827" type="marginalia">
|
||||
<pc:Coords points="208,1112 207,1113 172,1113 172,1114 172,1115 149,1115 148,1116 133,1116 133,1118 132,1119 132,1172 132,1172 132,1181 132,1182 132,1193 133,1194 134,1194 135,1195 135,1196 142,1196 143,1196 143,1197 194,1197 194,1196 195,1196 198,1196 198,1195 199,1194 204,1194 204,1193 205,1193 205,1188 206,1188 207,1188 207,1191 208,1191 208,1192 216,1192 216,1191 221,1191 222,1190 222,1188 223,1188 223,1180 222,1179 223,1178 224,1178 224,1170 224,1169 225,1169 225,1168 227,1167 228,1167 228,1166 231,1166 231,1164 232,1164 240,1164 240,1163 242,1163 242,1160 243,1160 243,1156 244,1156 244,1155 244,1154 245,1154 245,1152 246,1151 246,1137 245,1136 245,1124 244,1124 244,1124 244,1115 243,1114 243,1113 240,1113 240,1112 208,1112"/>
|
||||
<pc:TextLine id="region_0017_line_0001">
|
||||
<pc:Coords points="210,1110 206,1112 175,1111 170,1116 171,1114 152,1112 148,1115 136,1113 131,1120 132,1118 130,1136 132,1140 156,1142 153,1141 191,1143 188,1142 204,1144 208,1141 212,1143 216,1140 220,1142 224,1140 225,1141 225,1141 229,1137 227,1139 224,1138 236,1140 240,1137 238,1139 242,1136 240,1138 244,1136 245,1132 244,1134 246,1127 240,1122 243,1123 244,1117 242,1113 244,1116 236,1111 239,1112 210,1110"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0017_line_0002">
|
||||
<pc:Coords points="241,1135 237,1137 237,1136 233,1138 232,1136 228,1139 226,1137 222,1140 217,1138 213,1140 208,1139 204,1141 140,1140 142,1140 135,1139 130,1164 159,1168 156,1168 167,1169 164,1168 176,1170 174,1169 187,1171 184,1170 202,1172 206,1169 210,1171 214,1168 221,1170 226,1164 226,1166 224,1168 228,1165 227,1167 232,1160 231,1163 236,1164 240,1162 238,1164 243,1156 242,1159 244,1152 243,1155 244,1151 244,1153 241,1155 246,1148 245,1150 247,1140 241,1135"/>
|
||||
</pc:TextLine>
|
||||
<pc:TextLine id="region_0017_line_0003">
|
||||
<pc:Coords points="135,1165 130,1174 131,1172 129,1177 132,1181 130,1189 132,1193 136,1195 134,1194 140,1196 137,1195 150,1196 148,1196 188,1197 192,1195 191,1196 195,1194 196,1189 196,1192 197,1176 195,1172 196,1173 190,1168 192,1169 179,1168 181,1168 167,1167 169,1168 160,1166 163,1167 135,1165"/>
|
||||
</pc:TextLine>
|
||||
</pc:TextRegion>
|
||||
</pc:Page>
|
||||
</pc:PcGts>
|
||||
|
|
@ -1,16 +0,0 @@
|
|||
from eynollah.model_zoo import EynollahModelZoo
|
||||
|
||||
def test_trocr1(
|
||||
model_dir,
|
||||
):
|
||||
model_zoo = EynollahModelZoo(model_dir)
|
||||
try:
|
||||
from transformers import TrOCRProcessor, VisionEncoderDecoderModel
|
||||
model_zoo.load_model('trocr_processor')
|
||||
proc = model_zoo.get('trocr_processor', TrOCRProcessor)
|
||||
assert isinstance(proc, TrOCRProcessor)
|
||||
model_zoo.load_model('ocr', 'tr')
|
||||
model = model_zoo.get('ocr', VisionEncoderDecoderModel)
|
||||
assert isinstance(model, VisionEncoderDecoderModel)
|
||||
except ImportError:
|
||||
pass
|
||||
132
tests/test_run.py
Normal file
132
tests/test_run.py
Normal file
|
|
@ -0,0 +1,132 @@
|
|||
from os import environ
|
||||
from pathlib import Path
|
||||
import logging
|
||||
from PIL import Image
|
||||
from eynollah.cli import layout as layout_cli, binarization as binarization_cli
|
||||
from click.testing import CliRunner
|
||||
from ocrd_modelfactory import page_from_file
|
||||
from ocrd_models.constants import NAMESPACES as NS
|
||||
|
||||
testdir = Path(__file__).parent.resolve()
|
||||
|
||||
EYNOLLAH_MODELS = environ.get('EYNOLLAH_MODELS', str(testdir.joinpath('..', 'models_eynollah').resolve()))
|
||||
SBBBIN_MODELS = environ.get('SBBBIN_MODELS', str(testdir.joinpath('..', 'default-2021-03-09').resolve()))
|
||||
|
||||
def test_run_eynollah_layout_filename(tmp_path, subtests, pytestconfig, caplog):
|
||||
infile = testdir.joinpath('resources/kant_aufklaerung_1784_0020.tif')
|
||||
outfile = tmp_path / 'kant_aufklaerung_1784_0020.xml'
|
||||
args = [
|
||||
'-m', EYNOLLAH_MODELS,
|
||||
'-i', str(infile),
|
||||
'-o', str(outfile.parent),
|
||||
# subtests write to same location
|
||||
'--overwrite',
|
||||
]
|
||||
if pytestconfig.getoption('verbose') > 0:
|
||||
args.extend(['-l', 'DEBUG'])
|
||||
caplog.set_level(logging.INFO)
|
||||
def only_eynollah(logrec):
|
||||
return logrec.name == 'eynollah'
|
||||
runner = CliRunner()
|
||||
for options in [
|
||||
[], # defaults
|
||||
["--allow_scaling", "--curved-line"],
|
||||
["--allow_scaling", "--curved-line", "--full-layout"],
|
||||
["--allow_scaling", "--curved-line", "--full-layout", "--reading_order_machine_based"],
|
||||
["--allow_scaling", "--curved-line", "--full-layout", "--reading_order_machine_based",
|
||||
"--textline_light", "--light_version"],
|
||||
# -ep ...
|
||||
# -eoi ...
|
||||
# --do_ocr
|
||||
# --skip_layout_and_reading_order
|
||||
]:
|
||||
with subtests.test(#msg="test CLI",
|
||||
options=options):
|
||||
with caplog.filtering(only_eynollah):
|
||||
result = runner.invoke(layout_cli, args + options, catch_exceptions=False)
|
||||
print(result)
|
||||
assert result.exit_code == 0
|
||||
logmsgs = [logrec.message for logrec in caplog.records]
|
||||
assert str(infile) in logmsgs
|
||||
assert outfile.exists()
|
||||
tree = page_from_file(str(outfile)).etree
|
||||
regions = tree.xpath("//page:TextRegion", namespaces=NS)
|
||||
assert len(regions) >= 2, "result is inaccurate"
|
||||
regions = tree.xpath("//page:SeparatorRegion", namespaces=NS)
|
||||
assert len(regions) >= 2, "result is inaccurate"
|
||||
lines = tree.xpath("//page:TextLine", namespaces=NS)
|
||||
assert len(lines) == 31, "result is inaccurate" # 29 paragraph lines, 1 page and 1 catch-word line
|
||||
|
||||
def test_run_eynollah_layout_directory(tmp_path, pytestconfig, caplog):
|
||||
indir = testdir.joinpath('resources')
|
||||
outdir = tmp_path
|
||||
args = [
|
||||
'-m', EYNOLLAH_MODELS,
|
||||
'-di', str(indir),
|
||||
'-o', str(outdir),
|
||||
]
|
||||
if pytestconfig.getoption('verbose') > 0:
|
||||
args.extend(['-l', 'DEBUG'])
|
||||
caplog.set_level(logging.INFO)
|
||||
def only_eynollah(logrec):
|
||||
return logrec.name == 'eynollah'
|
||||
runner = CliRunner()
|
||||
with caplog.filtering(only_eynollah):
|
||||
result = runner.invoke(layout_cli, args)
|
||||
print(result)
|
||||
assert result.exit_code == 0
|
||||
logmsgs = [logrec.message for logrec in caplog.records]
|
||||
assert len([logmsg for logmsg in logmsgs if logmsg.startswith('Job done in')]) == 2
|
||||
assert any(logmsg for logmsg in logmsgs if logmsg.startswith('All jobs done in'))
|
||||
assert len(list(outdir.iterdir())) == 2
|
||||
|
||||
def test_run_eynollah_binarization_filename(tmp_path, subtests, pytestconfig, caplog):
|
||||
infile = testdir.joinpath('resources/kant_aufklaerung_1784_0020.tif')
|
||||
outfile = tmp_path.joinpath('kant_aufklaerung_1784_0020.png')
|
||||
args = [
|
||||
'-m', SBBBIN_MODELS,
|
||||
str(infile),
|
||||
str(outfile),
|
||||
]
|
||||
caplog.set_level(logging.INFO)
|
||||
def only_eynollah(logrec):
|
||||
return logrec.name == 'SbbBinarizer'
|
||||
runner = CliRunner()
|
||||
for options in [
|
||||
[], # defaults
|
||||
["--no-patches"],
|
||||
]:
|
||||
with subtests.test(#msg="test CLI",
|
||||
options=options):
|
||||
with caplog.filtering(only_eynollah):
|
||||
result = runner.invoke(binarization_cli, args + options)
|
||||
print(result)
|
||||
assert result.exit_code == 0
|
||||
logmsgs = [logrec.message for logrec in caplog.records]
|
||||
assert any(True for logmsg in logmsgs if logmsg.startswith('Predicting'))
|
||||
assert outfile.exists()
|
||||
with Image.open(infile) as original_img:
|
||||
original_size = original_img.size
|
||||
with Image.open(outfile) as binarized_img:
|
||||
binarized_size = binarized_img.size
|
||||
assert original_size == binarized_size
|
||||
|
||||
def test_run_eynollah_binarization_directory(tmp_path, subtests, pytestconfig, caplog):
|
||||
indir = testdir.joinpath('resources')
|
||||
outdir = tmp_path
|
||||
args = [
|
||||
'-m', SBBBIN_MODELS,
|
||||
'-di', str(indir),
|
||||
'-do', str(outdir),
|
||||
]
|
||||
caplog.set_level(logging.INFO)
|
||||
def only_eynollah(logrec):
|
||||
return logrec.name == 'SbbBinarizer'
|
||||
runner = CliRunner()
|
||||
with caplog.filtering(only_eynollah):
|
||||
result = runner.invoke(binarization_cli, args)
|
||||
print(result)
|
||||
assert result.exit_code == 0
|
||||
logmsgs = [logrec.message for logrec in caplog.records]
|
||||
assert len([logmsg for logmsg in logmsgs if logmsg.startswith('Predicting')]) == 2
|
||||
assert len(list(outdir.iterdir())) == 2
|
||||
|
|
@ -2,5 +2,6 @@ def test_utils_import():
|
|||
import eynollah.utils
|
||||
import eynollah.utils.contour
|
||||
import eynollah.utils.drop_capitals
|
||||
import eynollah.utils.drop_capitals
|
||||
import eynollah.utils.is_nan
|
||||
import eynollah.utils.rotate
|
||||
|
|
|
|||
|
|
@ -1,29 +0,0 @@
|
|||
# Use NVIDIA base image
|
||||
FROM nvidia/cuda:11.8.0-cudnn8-devel-ubuntu20.04
|
||||
|
||||
# Set the working directory
|
||||
WORKDIR /app
|
||||
|
||||
|
||||
# Set environment variable for GitPython
|
||||
ENV GIT_PYTHON_REFRESH=quiet
|
||||
|
||||
# Install Python and pip
|
||||
RUN apt-get update && apt-get install -y --fix-broken && \
|
||||
apt-get install -y \
|
||||
python3 \
|
||||
python3-pip \
|
||||
python3-distutils \
|
||||
python3-setuptools \
|
||||
python3-wheel && \
|
||||
rm -rf /var/lib/apt/lists/*
|
||||
|
||||
# Copy and install Python dependencies
|
||||
COPY requirements.txt .
|
||||
RUN pip install --no-cache-dir -r requirements.txt
|
||||
|
||||
# Copy the rest of the application
|
||||
COPY . .
|
||||
|
||||
# Specify the entry point
|
||||
CMD ["python3", "train.py", "with", "config_params_docker.json"]
|
||||
|
|
@ -1,83 +0,0 @@
|
|||
{
|
||||
"backbone_type" : "transformer",
|
||||
"task": "cnn-rnn-ocr",
|
||||
"n_classes" : 2,
|
||||
"max_len": 280,
|
||||
"n_epochs" : 3,
|
||||
"input_height" : 32,
|
||||
"input_width" : 512,
|
||||
"weight_decay" : 1e-6,
|
||||
"n_batch" : 4,
|
||||
"learning_rate": 1e-5,
|
||||
"save_interval": 1500,
|
||||
"patches" : false,
|
||||
"pretraining" : true,
|
||||
"augmentation" : true,
|
||||
"flip_aug" : false,
|
||||
"blur_aug" : true,
|
||||
"scaling" : false,
|
||||
"adding_rgb_background": true,
|
||||
"adding_rgb_foreground": true,
|
||||
"add_red_textlines": true,
|
||||
"white_noise_strap": true,
|
||||
"textline_right_in_depth": true,
|
||||
"textline_left_in_depth": true,
|
||||
"textline_up_in_depth": true,
|
||||
"textline_down_in_depth": true,
|
||||
"textline_right_in_depth_bin": true,
|
||||
"textline_left_in_depth_bin": true,
|
||||
"textline_up_in_depth_bin": true,
|
||||
"textline_down_in_depth_bin": true,
|
||||
"bin_deg": true,
|
||||
"textline_skewing": true,
|
||||
"textline_skewing_bin": true,
|
||||
"channels_shuffling": true,
|
||||
"degrading": true,
|
||||
"brightening": true,
|
||||
"binarization" : true,
|
||||
"pepper_aug": true,
|
||||
"pepper_bin_aug": true,
|
||||
"image_inversion": true,
|
||||
"scaling_bluring" : false,
|
||||
"scaling_binarization" : false,
|
||||
"scaling_flip" : false,
|
||||
"rotation": false,
|
||||
"color_padding_rotation": true,
|
||||
"padding_white": true,
|
||||
"rotation_not_90": true,
|
||||
"transformer_num_patches_xy": [56, 56],
|
||||
"transformer_patchsize_x": 4,
|
||||
"transformer_patchsize_y": 4,
|
||||
"transformer_projection_dim": 64,
|
||||
"transformer_mlp_head_units": [128, 64],
|
||||
"transformer_layers": 1,
|
||||
"transformer_num_heads": 1,
|
||||
"transformer_cnn_first": false,
|
||||
"blur_k" : ["blur","gauss","median"],
|
||||
"padd_colors" : ["white", "black"],
|
||||
"scales" : [0.6, 0.7, 0.8, 0.9],
|
||||
"brightness" : [1.3, 1.5, 1.7, 2],
|
||||
"degrade_scales" : [0.2, 0.4],
|
||||
"pepper_indexes": [0.01, 0.005],
|
||||
"skewing_amplitudes" : [5, 8],
|
||||
"flip_index" : [0, 1, -1],
|
||||
"shuffle_indexes" : [ [0,2,1], [1,2,0], [1,0,2] , [2,1,0]],
|
||||
"thetha" : [0.1, 0.2, -0.1, -0.2],
|
||||
"thetha_padd": [-0.6, -1, -1.4, -1.8, 0.6, 1, 1.4, 1.8],
|
||||
"white_padds" : [0.1, 0.3, 0.5, 0.7, 0.9],
|
||||
"number_of_backgrounds_per_image": 2,
|
||||
"continue_training": false,
|
||||
"index_start" : 0,
|
||||
"dir_of_start_model" : " ",
|
||||
"weighted_loss": false,
|
||||
"is_loss_soft_dice": false,
|
||||
"data_is_provided": false,
|
||||
"dir_train": "/home/vahid/extracted_lines/1919_bin/train",
|
||||
"dir_eval": "/home/vahid/Documents/test/sbb_pixelwise_segmentation/test_label/pageextractor_test/eval_new",
|
||||
"dir_output": "/home/vahid/extracted_lines/1919_bin/output",
|
||||
"dir_rgb_backgrounds": "/home/vahid/Documents/1_2_test_eynollah/set_rgb_background",
|
||||
"dir_rgb_foregrounds": "/home/vahid/Documents/1_2_test_eynollah/out_set_rgb_foreground",
|
||||
"dir_img_bin": "/home/vahid/extracted_lines/1919_bin/images_bin",
|
||||
"characters_txt_file":"/home/vahid/Downloads/models_eynollah/model_eynollah_ocr_cnnrnn_20250930/characters_org.txt"
|
||||
|
||||
}
|
||||
|
|
@ -1,54 +0,0 @@
|
|||
{
|
||||
"backbone_type" : "nontransformer",
|
||||
"task": "segmentation",
|
||||
"n_classes" : 3,
|
||||
"n_epochs" : 1,
|
||||
"input_height" : 672,
|
||||
"input_width" : 448,
|
||||
"weight_decay" : 1e-6,
|
||||
"n_batch" : 4,
|
||||
"learning_rate": 1e-4,
|
||||
"patches" : false,
|
||||
"pretraining" : true,
|
||||
"augmentation" : false,
|
||||
"flip_aug" : false,
|
||||
"blur_aug" : true,
|
||||
"scaling" : true,
|
||||
"adding_rgb_background": false,
|
||||
"adding_rgb_foreground": false,
|
||||
"add_red_textlines": false,
|
||||
"channels_shuffling": true,
|
||||
"degrading": true,
|
||||
"brightening": true,
|
||||
"binarization" : false,
|
||||
"scaling_bluring" : false,
|
||||
"scaling_binarization" : false,
|
||||
"scaling_flip" : false,
|
||||
"rotation": false,
|
||||
"rotation_not_90": true,
|
||||
"transformer_num_patches_xy": [14, 21],
|
||||
"transformer_patchsize_x": 1,
|
||||
"transformer_patchsize_y": 1,
|
||||
"transformer_projection_dim": 64,
|
||||
"transformer_mlp_head_units": [128, 64],
|
||||
"transformer_layers": 1,
|
||||
"transformer_num_heads": 1,
|
||||
"transformer_cnn_first": true,
|
||||
"blur_k" : ["blur","gauss","median"],
|
||||
"scales" : [0.6, 0.7, 0.8, 0.9],
|
||||
"brightness" : [1.3, 1.5, 1.7, 2],
|
||||
"degrade_scales" : [0.2, 0.4],
|
||||
"flip_index" : [0, 1, -1],
|
||||
"shuffle_indexes" : [ [0,2,1], [1,2,0], [1,0,2] , [2,1,0]],
|
||||
"thetha" : [5, -5],
|
||||
"number_of_backgrounds_per_image": 2,
|
||||
"continue_training": false,
|
||||
"index_start" : 0,
|
||||
"dir_of_start_model" : " ",
|
||||
"weighted_loss": false,
|
||||
"is_loss_soft_dice": true,
|
||||
"data_is_provided": false,
|
||||
"dir_train": "/entry_point_dir/train",
|
||||
"dir_eval": "/entry_point_dir/eval",
|
||||
"dir_output": "/entry_point_dir/output"
|
||||
}
|
||||
|
|
@ -1,8 +0,0 @@
|
|||
{
|
||||
"use_case": "textline",
|
||||
"textregions":{ "rest_as_paragraph": 1, "header":2 , "heading":2 , "marginalia":3 },
|
||||
"imageregion":4,
|
||||
"separatorregion":5,
|
||||
"graphicregions" :{"rest_as_decoration":6},
|
||||
"columns_width":{"1":1000, "2":1300, "3":1600, "4":2000, "5":2300, "6":2500}
|
||||
}
|
||||
|
|
@ -1,6 +0,0 @@
|
|||
sacred
|
||||
seaborn
|
||||
numpy <1.24.0
|
||||
tqdm
|
||||
imutils
|
||||
scipy
|
||||
|
|
@ -1,3 +0,0 @@
|
|||
{
|
||||
"scales" : [ 0.5, 0.55, 0.6, 0.65, 0.7, 0.75, 0.8, 0.85, 0.9]
|
||||
}
|
||||
Loading…
Add table
Add a link
Reference in a new issue